 遗传多样性分析
遗传多样性分析
 (Liza affinis)是分布于西北太平洋的日本南部及中国台湾海域等的经济鱼类。为了解中国前鳞
(Liza affinis)是分布于西北太平洋的日本南部及中国台湾海域等的经济鱼类。为了解中国前鳞 的遗传背景,该研究分析了采自中国4省82尾样本COⅠ基因5'端712 bp序列,共发现21个单倍型,遗传多样性(Hd=0.4840±0.0700, π=0.0010±0.0002)较低。根据群体所属的地区、海域、海峡进行分组,通过AMOVA分析,得出组间的Fst值皆小于0.05,P值皆大于0.05,组间变异的比例极低(–0.6%~0.46%),表明中国前鳞
的遗传背景,该研究分析了采自中国4省82尾样本COⅠ基因5'端712 bp序列,共发现21个单倍型,遗传多样性(Hd=0.4840±0.0700, π=0.0010±0.0002)较低。根据群体所属的地区、海域、海峡进行分组,通过AMOVA分析,得出组间的Fst值皆小于0.05,P值皆大于0.05,组间变异的比例极低(–0.6%~0.46%),表明中国前鳞 群体间无明显分化,其原因可能是:1)频繁的基因交流。前鳞
群体间无明显分化,其原因可能是:1)频繁的基因交流。前鳞 的洄游范围较大,且受海流影响,导致不同地区的群体间有着密切的基因交流;2)近期种群扩张事件。中国前鳞
的洄游范围较大,且受海流影响,导致不同地区的群体间有着密切的基因交流;2)近期种群扩张事件。中国前鳞 整体的Fu’s Fs值为显著性负值(FS= –20.3900,P=0),核苷酸错配峰图为明显单峰,单倍型网络图呈星状结构,均表明中国前鳞
整体的Fu’s Fs值为显著性负值(FS= –20.3900,P=0),核苷酸错配峰图为明显单峰,单倍型网络图呈星状结构,均表明中国前鳞 群体经历过种群扩张,估算扩张大约发生在13.4199~1.4911万年前,可能是冰期和间冰期的交替中海平面的变化所致。此外,洞头群体的遗传多样性(Hd=0.8080±0.1130,π=0.0021±0.0006)明显高于其他地理群体,建议将其作为优先保护的对象。
群体经历过种群扩张,估算扩张大约发生在13.4199~1.4911万年前,可能是冰期和间冰期的交替中海平面的变化所致。此外,洞头群体的遗传多样性(Hd=0.8080±0.1130,π=0.0021±0.0006)明显高于其他地理群体,建议将其作为优先保护的对象。 遗传多样性 COⅠ基因 中国沿海
遗传多样性 COⅠ基因 中国沿海 遗传多样性是物种生存与进化的物质基础,开展鱼类遗传多样性研究可以为遗传育种和种质资源保护提供重要参考(Ward, 2000)。动物线粒体属于母系遗传,无遗传物质的重组,且其进化速度比核基因快,较少的样本就可较好地反映该种群的遗传结构,是动物遗传多样性研究中理想的分子标记(Xu et al, 2014)。而且,动物线粒体的COⅠ序列具有一定的进化变异,可以较好地反映同种生物的不同群体间的遗传差异,除了被广泛应用于DNA条形码的研究中,也适用于遗传多样性的分析研究(柳淑芳等, 2016; Chang et al, 2016; Xu et al, 2012; 李大命等, 2017)。
前鳞




为了研究中国前鳞

运用MEGA 7.0(Kumar et al, 2016)对测序峰图进行人工校对,并计算碱基组成、颠换率。通过DnaSP 5.10计算单倍型数、单倍型多样性(Hd)、核苷酸多样性(π) (Librado et al, 2009)。使用Network (Bandelt et al, 1999)分析单倍型的分布情况,通过Arlequin 3.5进行AMOVA分析、核苷酸错配分析、Tajima's D和Fu's Fs中性检验,计算群组间的Fst值、SSD值、Rg值、τ值等参数(Excoffier et al, 2010)。运用SAMOVA 2.0进行空间分子变异分析。根据公式[T=(τ/2μk)×代时]计算种群扩张时间,其中,τ、μ、k、T依次代表种群扩张时间参数、序列的变异速率[线粒体COⅠ基因进化速率为(1%~3%)/百万年(Wang et al, 2013)]、序列长度以及种群扩张时间(Rogers et al, 1992)。





为了方便比较分布在不同地理位置的前鳞





在全长为712 bp的序列中,没有发现碱基的插入和缺失,碱基A、T、C和G的含量分别为23.5%、29.4%、28.8%和18.3%,A+T含量(52.9%)略高于C+G含量(47.1%),与其他硬骨鱼类COⅠ序列碱基组成特征基本一致。其中,共有18个多态位点、6个简约信息位点,颠换与转换的比值为4.7,表明序列的突变尚未达到饱和,适合进行系统发育分析(Kumar, 2005)。此外,还发现21个单倍型,总体遗传多样性偏低(Hd=0.4840±0.0700,π=0.0010±0.0002),其中,洞头群体最高,岱山群体最低,东海群体的遗传多样性高于南海群体,具体的遗传多样性情况见表 1。
|
|
表 1 中国沿海前鳞 |
单倍型最大简约性网络图(图 1)呈现星状结构,其中,Hap1的数量最多(59),由全部群体共享。Hap3的数量为3,由东兴群体与平潭群体共享。Hap11的数量为2,由平潭群体与洞头群体共享,其余均为各个群体特有的单倍型。也就是说,不同地理来源的个体相互聚集分布,不同的地区中存在相同的单倍型,表明前鳞
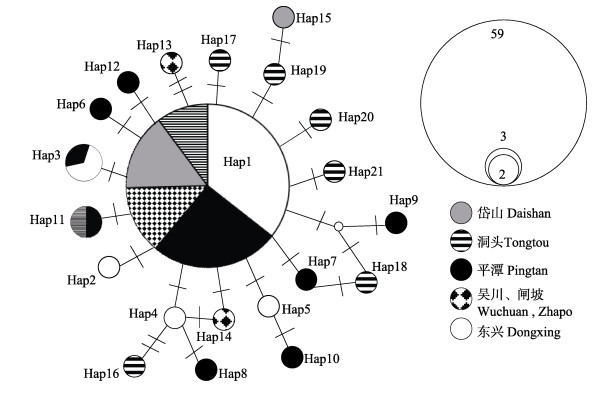
|
图 1 前鳞 |
在AMOVA分析(表 2)中,按照上文的群体划分进行分析:东兴、平潭、闸坡、广东、洞头等5个群体间的Fst=-0.0221~0.0141(P > 0.05),组间的遗传变异比例达-0.26%。东海群体与南海群体间的Fst= 0.0020 (P > 0.05),组间的遗传变异比例达0.46%。东兴、广东、东海3个群体间的Fst= -0.0159~0.0141 (P > 0.05),组间的遗传变异比例为-0.6%。上述各类划分中,群体间的Fst值皆小于0.05,均表现出不显著水平(P > 0.05),且组间的遗传变异比例均属于极低水平,表明中国前鳞
|
|
表 2 前鳞 |

中性检验的分析结果(表 3)如下:Tajima's D方面,洞头、平潭、广东、东兴4个群体均为显著性负值,其余为非显著性负值。Fu's Fs方面,洞头、广东、东兴3个群体均呈显著性负值,总体也是显著性负值(FS=-20.3900,P=0)。Ramos-Onsins & Rozas's R2检验的分析显示(表 3):R2的范围是0.1610~0.1620 (P < 0.05),SSD值(0.0010~0.0220)和Rg值(0.0500~ 0.7200)均较小,且多数呈不显著性(P > 0.05),表明中国前鳞


|
|
表 3 前鳞 |
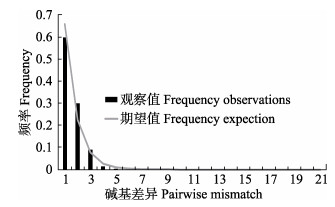
|
图 2 前鳞 |

某物种的遗传多样性丰富与否和该物种所反映的单倍型多样性(Hd)及核苷酸多样性(π)密切相关(Bonin et al, 2010)。该研究中,中国前鳞











该研究中AMOVA分析所得的组间变异比例(–0.6%~0.46%)与许则滩(2015)对浙江沿岸前鳞












鱼类作为水生生态系统的高级消费者,处于食物链的较高位置,是生态系统中物质循环、能量流动的重要参与者,与水生环境有着密切的联系,故各种水体污染和不适当的人类活动都可能对鱼类的生存产生不利的影响。而正常的生态系统功能的维持,需要制定相关的种质资源保护方案。仅就该研究中,中国前鳞



Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 1999, 16(1): 37-48 |
Bonin A, Nicole F, Pompanon F, et al. Population adaptive index: A new method to help measure intraspecific genetic diversity and prioritize populations for conservation. Conservation Biology, 2010, 21(3): 697-708 |
Cao Y, Zhang Q, Gong YY, et al. Genetic variation of Scomberomorus niphonius in the coastal waters of China based on mtDNA COⅠsequences. Marine Fisheries, 2015, 37(6): 485-493 |
曹艳, 章群, 宫亚运, 等. 基于线粒体COⅠ序列的中国沿海蓝点马鲛遗传多样性. 海洋渔业, 2015, 37(6): 485-493 DOI:10.3969/j.issn.1004-2490.2015.06.001 |
Chang CH, Lin HY, Ren Q, et al. DNA barcode identification of fish products in Taiwan: Government-commissioned authentication cases. Food Control, 2016, 66: 38-43 DOI:10.1016/j.foodcont.2016.01.034 |
Excoffier L, Lischer H. Arlequin suite Ver 3. 5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 2010, 10(3): 564-567 DOI:10.1111/men.2010.10.issue-3 |
Ju HL. Studies on the decrement of fishery resource in the South China Sea. Southeast Asian Studies, 2012(6): 51-55 |
鞠海龙. 南海渔业资源衰减相关问题研究. 东南亚研究, 2012(6): 51-55 DOI:10.3969/j.issn.1008-6099.2012.06.007 |
Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis Version 7. 0 for bigger datasets. Molecular Biology and Evolution, 2016, 33(7): 54-58 |
Kumar S. Molecular clocks: Four decades of evolution. Nature Reviews Genetics, 2005, 6(8): 654-662 DOI:10.1038/nrg1659 |
Le XL, Zhang Q, Zhao S, et al. A fast and efficient method for isolation of genomic DNA from fish specimens. Biotechnology Bulletin, 2010, 20(2): 202-204 |
乐小亮, 章群, 赵爽, 等. 一种高效快速的鱼类标本基因组DNA提取方法. 生物技术通报, 2010, 20(2): 202-204 |
Lesica P, Allendorf FW. When are peripheral populations valuable for conservation?. Conservation Biology, 1995, 9(4): 753-760 DOI:10.1046/j.1523-1739.1995.09040753.x |
Li DM, Zhang TQ, Tang SK, et al. Genetic variation in two phenotypical populations of Corbicula fluminea in Hongze Lake. Progress in Fishery Sciences, 2017, 38(4): 111-117 |
李大命, 张彤晴, 唐晟凯, 等. 洪泽湖河蚬(Corbicula fluminea)2种表型群体的遗传变异分析. 渔业科学进展, 2017, 38(4): 111-117 |
Li S. Population genetics studies on four species of Crassostrea oysters. Masterxs Thesis of Ocean University of China, 2015: 1-103
|
李双. 四种巨蛎属牡蛎群体遗传学研究. 中国海洋大学硕士研究生学位论文, 2015: 1-103
|
Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 2009, 25(11): 1451-1452 DOI:10.1093/bioinformatics/btp187 |
Lin HJ, Shao KT, Kuo SR, et al. A trophic model of a sandy barrier lagoon at Chiku in Southwestern Taiwan. Estuarine Coastal and Shelf Science, 1999, 48(5): 575-588 DOI:10.1006/ecss.1998.0457 |
Liu JQ, Wang WY. Quaternary geological dating and time scale. Quaternary Sciences, 1997, 17(3): 193-202 |
刘嘉麒, 王文远. 第四纪地质定年与地质年表. 第四纪研究, 1997, 17(3): 193-202 DOI:10.3321/j.issn:1001-7410.1997.03.001 |
Liu SF, Li XR, Li D, et al. Development of DNA barcode-microarray for identification of Engraulidae fishes. Progress in Fishery Sciences, 2016, 37(6): 19-25 |
柳淑芳, 李献儒, 李达, 等. 鳀科(Engraulidae)鱼类DNA条形码电子芯片研究. 渔业科学进展, 2016, 37(6): 19-25 |
Lu ZK. Studies on the reproductive ecology of Mugilidae. Journal of Zhejiang Ocean University, 1985, 4(1): 67-72 |
陆忠康. 关于鲻科鱼类繁殖生态学的研究. 浙江海洋学院学报, 1985, 4(1): 67-72 |
Meng W, Gao TX, Song L, et al. almost periodic solution of ordinary differential equations. Periodical of Ocean University of China, 2007, 37(supp): 181-184 |
孟玮, 高天翔, 宋林, 等. 三种鲻科鱼的同工酶分析及鉴别研究. 中国海洋大学学报, 2007, 37(supp): 181-184 |
Nakajima K, Kitada S, Habara Y, et al. Genetic effects of marine stock enhancement: A case study based on the highly piscivorous Japanese spanish mackerel. Canadian Journal of Fisheries and Aquatic Sciences, 2014, 71(71): 301-314 |
Ni G, Li QI, Kong L, et al. Comparative phylogeography in marginal seas of the northwestern Pacific. Molecular Ecology, 2014, 23(3): 534-548 |
Qian GY. Sex and artificial control of fish. Chinese Journal of Zoology, 2000, 35(1): 47-52 |
钱国英. 鱼类的性别及人工控制. 动物学杂志, 2000, 35(1): 47-52 DOI:10.3969/j.issn.0250-3263.2000.01.019 |
Richardson N, Gordon AK, Muller WJ, et al. A weight-of- evidence approach to determine estuarine fish health using indicators from multiple levels of biological organization. Aquatic Conservation Marine & Freshwater Ecosystems, 2011, 21(5): 423-432 |
Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution, 1992, 9(3): 52-69 |
Shi Q, Fan MJ, Zhang Y. Economically important fishes in China. Wuhan: Huazhong University of Science and Technology Press, 2015: 360-363
|
石琼, 范明君, 张勇. 中国经济鱼类志. 武汉: 华中科技大学出版社, 2015: 360-363
|
Stirling CH, Esat TM, Mcculloch MT, et al. High-precision U-series dating of corals from Western Australia and implications for the timing and duration of the last interglacial. Earth & Planetary Science Letters, 1995, 135(1): 115-130 |
Sun DR, Chen Z. Fish index in the South China Sea(Volume Ⅰ). Beijing: China Ocean Press, 2013: 375-377
|
孙典荣, 陈铮. 南海鱼类检索(上册). 北京: 海洋出版社, 2013: 375-377
|
Wang L, Shi X, Su Y, et al. Genetic divergence and historical demography in the endangered large yellow croaker revealed by mtDNA. Biochemical Systematics and Ecology, 2013, 46(46): 137-144 |
Wang P. Response of Western Pacific marginal seas to glacial cycles: Paleoceanographic and sedimentological features 1. Marine Geology, 1999, 156(1-4): 5-39 DOI:10.1016/S0025-3227(98)00172-8 |
Ward RD. Genetics in fisheries management. Hydrobiologia, 2000, 420(1): 191-201 DOI:10.1023/A:1003928327503 |
Winograd IJ, Landwehr JM, Ludwig KR, et al. Duration and structure of the past four interglaciations. Quaternary Research, 1997, 48(2): 141-154 DOI:10.1006/qres.1997.1918 |
Xu D, Lou B, Shi H, et al. Genetic diversity and population structure of Nibea albiflora, in the China Sea revealed by mitochondrial COⅠsequences. Biochemical Systematics and Ecology, 2012, 45(12): 158-165 |
Xu H, Zhang Y, Xu D, et al. Genetic population structure of miiuy croaker (Miichthys miiuy) in the yellow and east china seas base on mitochondrial COⅠsequences. Biochemical Systematics & Ecology, 2014, 54(1): 240-246 |
Xu ZT. Molecular identification and phylogeny of larval mugilids in Zhejiang coastal sea. Masterxs Thesis of Zhejiang Ocean University, 2015: 1-68
|
许则滩. 浙江沿岸海域幼鱼期鲻科鱼类的分子鉴定和系统发育研究. 浙江海洋学院硕士研究生学位论文, 2015: 1-68
|
Yang XS, Zhang Q, Yu FY, et al. MtDNA ND2 sequence-based genetic analysis of Anabas testudineus from South China and Lancang/Meko. South China Fisheries Science, 2017, 13(3): 43-50 |
杨喜书, 章群, 余帆洋, 等. 华南6水系与澜沧江-湄公河攀鲈线粒体ND2基因的遗传多样性分析. 南方水产科学, 2017, 13(3): 43-50 DOI:10.3969/j.issn.2095-0780.2017.03.006 |



