2. 上海海洋大学 水产科学国家级实验教学示范中心 上海 201306
2. National Demonstration Center for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai 201306
几丁质(Chitin)是昆虫和甲壳动物外骨骼和围食膜的主要分子成分,是由β-1, 4糖苷键连接的N-乙酰- β-D-氨基葡萄糖直链多聚糖,其含量仅次于纤维素,为第二大天然多糖(Zhang et al, 2013; Qiang et al, 2018)。几丁质酶(Chitinase)是广泛存在于动物、植物、真菌、细菌和病毒中的一种能够随机催化几丁质聚合物水解的酶(Adrangi et al, 2013; Umemoto et al, 2013)。在甲壳动物中,几丁质酶在几丁质类食物的消化、蜕皮和应激反应等生理功能中起关键作用(Zou et al, 2004; Merzendorfer, 2006; Gao et al, 2017)。
几丁质酶基因是一个具有GH18和GH19两大家族的多基因家族(Gao et al, 2018; Gayathri et al, 2018),目前,在隶属于GH18家族的主要经济甲壳动物中均已开展了几丁质酶基因相关功能的研究,并且根据基因结构特征将其归为7大类(Group1~7,其所属的基因也大多相应的命名为Cht1~7),但三疣梭子蟹(Portunus trituberculatus)几丁质酶基因仅发现了其中3类,包括Group3(张凤等, 2017)、Group5(宋柳等, 2019)和Group7(张凤等, 2015),还有很多未知的基因有待发掘。甲壳动物Group1几丁质酶基因多在肝胰腺中有较高表达,而肝胰腺是消化酶分泌的重要器官,表明其发挥着消化几丁质类食物的功能。Salma等(2012)在日本仿长额虾(Pandalopsis japonica)研究中发现,Pj-Cht1主要在肝胰腺中表达,可能在几丁质类食物的消化中起作用。凡纳滨对虾(Litopenaeus vannamei) LvChi1基因(Huang et al, 2010; Rocha et al, 2012)和拟穴青蟹(Scylla paramamosain) (Zhou et al, 2018) SpCht1基因均在肝胰腺中特异性表达,推测其参与了几丁质类食物的消化。类似结果在中华绒螯蟹(Eriocheir sinensis) (Li et al, 2015)、日本对虾(Marsupenaeus japonicus) (Watanabe et al, 1996、1998)、斑节对虾(Penaeus monodon) (Proespraiwong et al, 2010)和中国对虾(Fenneropenaeus chinenis)(Priya et al, 2009)等物种中也有发现。此外,也有研究显示,在尾扇、眼柄、鳃等组织(Watanabe et al, 1997; Rocha et al, 2012)亦有几丁质酶Group1基因的表达,暗示了其功能的多样性,可能参与了多种生理学进程。例如,日本沼虾(Macrobrachium nipponense) (Zhang et al, 2014) MnCht1A和MnCht1B除了在肝胰腺中高表达外,在表皮和肠道中亦有少量分布,表明该基因可能还在肠内围食膜中内源几丁质的降解以及几丁质外骨骼的分解中起作用。然而,Group1几丁质酶基因的结构及功能虽在其他虾蟹中有较多研究,但在三疣梭子蟹中相关的内容却鲜有报道。
本研究中,通过RACE克隆、生物信息学分析及qRT-PCR等方法对三疣梭子蟹PtCht1基因的结构及功能进行探究,明确基因的分类,解析PtCht1在促进消化等方面的功能,可为三疣梭子蟹几丁质酶GH18家族Group1基因的研究奠定基础,为耐低盐新品种的培育提供新思路。
1 材料与方法 1.1 实验材料实验材料取自中国水产科学研究院黄海水产研究所实验基地—山东潍坊昌邑市海丰水产有限公司养殖的健康三疣梭子蟹,湿重为(5.78±1.11) g。室内养蟹池4个,体积均为20 m3,每池150只蟹,暂养7 d。盐度33、pH 8.7的自然海水养殖,水温控制在(25±1)℃,保持氧气供应充足,每天定时更换1/3水量,每天18:00定时投喂新鲜蓝蛤(Potamocorbula laevis)。
1.2 全长cDNA的克隆及测序随机取8只(雌雄各4只)暂养后四肢健全且充满活力的健康三疣梭子蟹,解剖取样,包括心脏、肝胰腺、肠、鳃、胸神经节、肌肉、脑、眼柄、胃、表皮、精巢和卵巢,液氮冷冻后,将所有样品的同一组织混合后进行研磨,Trizol法提取总RNA,通过Thermo的NanoDrop 2000核酸定量仪与1%琼脂糖凝胶电泳检测其完整性,选取高质量的总RNA,混匀后用SMARTTM RACE Amplification Kit合成用于3′和5′ RACE的cDNA模板。依据三疣梭子蟹高通量测序数据库中PtCht1的已知表达序列标签(Expressed Sequence Tag, EST),运用Primer Premier 5.0软件设计3′和5′RACE特异性引物(PtCht1 F和PtCht1 R)、通用引物(UPM)和ORF验证引物(PtCht1 Fi和PtCht1 Ri)(表 1)。利用Advantage 2 PCR Kit对3′和5′末端进行扩增。纯化后的扩增产物与pMD18-T载体连接后转入DH5α大肠杆菌感受态细胞中扩大培养,挑取阳性单克隆,利用DNA测序通用引物M13-47/48进行菌落PCR鉴定,将目的产物送至睿博生物公司进行测序。利用Vector NTI 11.0去掉测序结果中冗余序列后,拼接获得完整的基因序列。
|
|
表 1 三疣梭子蟹PtCht1基因克隆和实时荧光定量所用引物序列 Tab.1 Primer sequences for cloning and qRT-PCR of PtCht1 gene in P. trituberculatus |
利用软件BioEdit预测ORF开放阅读框,并翻译蛋白序列; 将蛋白序列提交至在线分析软件SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/),预测信号肽; 利用NCBI的保守结构域(CDD)数据库(http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi)和Pfam数据库(http://pfam.sanger.ac.uk/)进行功能结构域的预测和确定; 使用DNAMAN分析软件将三疣梭子蟹的氨基酸序列与其他物种进行多重序列比对; 通过软件MEGA 6.0以Neighbor Joining法构建系统进化树(段亚飞等, 2013)。
1.4 蜕皮周期实验根据甲壳硬度和游泳足趾节末端新旧表皮的变化(沈洁等, 2011),将部分暂养7 d后的健康三疣梭子蟹分成蜕皮间期(甲壳硬,无新表皮出现)、蜕皮前期(甲壳硬,出现新表皮)和蜕皮后期(甲壳柔软),每个时期随机取3只,取其血细胞、第1对鳃、第6对鳃、胃、肝胰腺、心脏、表皮、肌肉、眼柄和肠组织于液氮速冻保存,实验设置3个平行。
1.5 盐度胁迫实验正式实验前,随机挑取240只暂养7 d后充满活力的健康三疣梭子蟹进行低盐胁迫预实验,实验方法参照隋延鸣等(2012)。设置4个盐度梯度,分别为33(对照)、13、11和9,每个盐度下放置20只蟹,设置3个平行,统计24、36、48和72 h三疣梭子蟹死亡数,计算结果显示,72 h半致死盐度为11,由此正式实验设置2个盐度,即对照组(33)和低盐组(11)。低盐度海水由自然海水(33)与淡水调配而成,每组设3个平行,每个平行50只蟹。分别在胁迫后0、3、6、12、24、48和72 h取3只梭子蟹的肝胰腺和鳃组织,液氮速冻保存,便于后续提取RNA。
1.6 PtCht1基因mRNA的实时定量PCR检测将获得的组织样品利用Trizol法提取总RNA,检测方法同1.2。选取高质量的总RNA样品,参照TOYOBO ReverTra Ace qPCR RT Master Mix with gDNA Remover试剂盒说明书,反转录成cDNA,用于后续的实时荧光定量PCR分析(qRT-PCR)。根据拼接获得的PtCht1基因cDNA序列全长和已知的三疣梭子蟹管家基因β-actin,利用软件Primer Premier 5.0设计荧光定量特异性引物(PtCht1 qF和PtCht1qR)和内参引物(β-actin-F和β-actin-R) (表 1),通过实时定量PCR法对PtCht1基因在各个组织和各实验组中的相对表达量进行检测分析,反应体系及程序参照TOYOBO SYBR® Green Real-time PCR Master Mix说明书进行,结果采用2-ΔΔCt方法计算,借助SPSS 19.0对数据进行单因素方差分析(One-way ANOVA),利用Origin Pro和Excel软件将结果整理统计,P < 0.05为差异显著。
2 结果 2.1 PtCht1基因的cDNA全长克隆及序列分析以三疣梭子蟹各组织的混合cDNA为模板进行RACE扩增,获得3′-981 bp和5′-477 bp的cDNA片段。将扩增的片段与已知的EST序列拼接,得到完整的几丁质酶1基因的cDNA,命名为PtCht1 (GenBank No.: KM100752)。基因结构如图 1所示,序列全长共2220 bp,其中,3′端及5′端非编码区分别为324 bp和144 bp,开放阅读框(Open Reading Frame, ORF)为1752 bp,共编码氨基酸583个,分子量为65.68 kDa,理论等电点为5.84。通过DNAstar 7.1.0 (Lasergene)软件包中EditSeq蛋白统计程序分析一级结构的基本性质。结果显示,PtCht1基因编码的氨基酸组成中强碱性(R和Lys)、强酸性(D和E)、疏水性(A、I、L、F、W和V)和极性氨基酸(S、C、N、Q、Y和T)残基的数目分别为78、83、266和141个。此外,图 1结合图 2蛋白结构域预测结果显示,N端存在22个氨基酸的信号肽序列,46~397个氨基酸为1个含4个保守基序的糖基水解酶第18家族催化结构域(GH18 catalytic domain)和活性位点161FDGFDL DWE169,在463~518个氨基酸处,含6个半胱氨酸(Cys)和几丁质结合结构域(ChtBD2)。
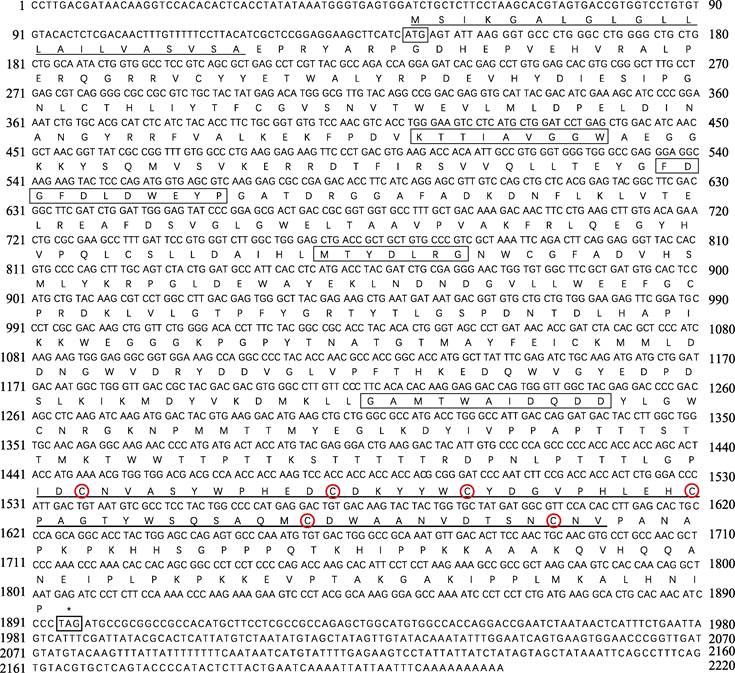
|
图 1 三疣梭子蟹PtCht1基因核苷酸序列及其推导的氨基酸序列 Fig.1 Nucleotide sequence and deduced amino acids sequence of P. trituberculatus PtCht1 起始密码子(ATG)和终止密码子(TAG)用细线方框标出; 信号肽以细下划线标出; 几丁质酶第18家族保守基序用粗线方框标出; ChtBD2结构域用粗下划线标出; 6个半胱氨酸用红色圆圈标出 The letters in the box indicated the start codon (ATG) and the stop codon (TAG). Signal peptide is underlined with thin line, the chitinase family 18 conserved motifs of catalytic domain are framed by black box, chitin-binding domain (ChtBD2) with thick line, six cysteines are marked with red circles |

|
图 2 三疣梭子蟹PtCht1基因编码的蛋白结构域位置 Fig.2 The protein domain architecture of P. trituberculatus PtCht1 |
将三疣梭子蟹Cht1的氨基酸序列与其他物种进行同源性比对分析(图 3)可知,PtCht1基因与拟穴青蟹SpCht1的同源性最高,为92.80%,与锯缘青蟹(Scylla serrata)、日本蟳(Charybdis japonica)、凡纳滨对虾、日本对虾、日本沼虾和中华绒螯蟹的同源性次之,分别为91.94%、90.85%、80.87%、76.98%、75.70%和75.00%。此外,甲壳动物几丁质酶GH18催化结构域的4个保守基序MotifⅠ~Ⅳ和含6个Cys的几丁质结合结构域ChtBD2在上述几个物种中均存在。系统进化树图谱显示(图 4),甲壳动物第18家族几丁质酶主要分为7大类,分别以Cht1、Cht2、Cht3、Cht4、Cht5、Cht6和Cht聚类于Group 1~7。三疣梭子蟹PtCht1基因与凡纳滨对虾、日本对虾、斑节对虾、锯缘青蟹、中国对虾、日本沼虾和日本仿长额虾的Cht1聚为一类,隶属于Group1。

|
图 3 三疣梭子蟹PtCht1氨基酸序列的多序列比对 Fig.3 Multiple alignment of the deduced amino acid sequences of P. trituberculatus PtCht1 with others 几丁质酶第18家族保守基序以红色方框表示,几丁质结合结构域(ChtBD2)中的6个半胱氨酸(Cys)以红色箭头表示Sp:拟穴青蟹; Ss:锯缘青蟹; Cj:日本蟳; Lv:凡纳滨对虾; Pj:日本对虾; Mn:日本沼虾; Es:中华绒螯蟹 The chitinase family 18 conserved motifs are framed by red box, the six cysteines (Cys) in the chitin binding domain (ChtBD2) are marked by red arrows Sp: Scylla paramamosain; Ss: Scylla serrata; Cj: Charybdis japonica; Lv: Litopenaeus vannamei; Pj: Marsupenaeus japonicus; Mn: Macrobrachium nipponense; Es: Eriocheir sinensis |
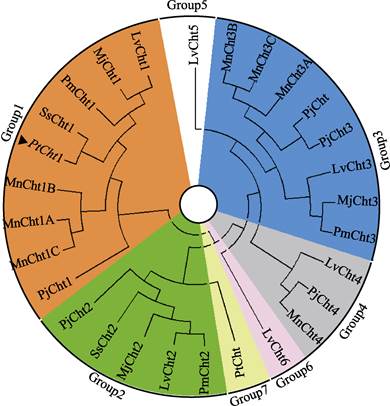
|
图 4 三疣梭子蟹几丁质酶蛋白PtCht1的系统进化树 Fig.4 Phylogenetic analysis of chitinase PtCht1 from P. trituberculatus ▲:三疣梭子蟹(Portunus trituberculatus)几丁质酶蛋白PtCht1;各物种几丁质酶GenBank登录号:日本对虾(Marsupenaeus japonicus)Cht1(BAA12287)、Cht2 (BAA14014)、Cht3(BAA22854);日本沼虾(Macrobrachium nipponense) Cht1A (KF466274.1)、Cht1B (KF466275.1)、Cht1C (AHL24866.1)、Cht3A (KF466276.1)、Cht3B (KF466277.1)、Cht3C (KF466278.1)、Cht4 (KF466279.1);斑节对虾(Penaeus monodon) Cht1 (AAD40313.1)、Cht2 (ADG22164.1)、Cht3 (ADG22163.1);锯缘青蟹(Scylla serrata) Cht (ABY85409.1)、Cht1 (ACG60512.1)、Cht2 (ACZ53950.1);凡纳滨对虾(Litopenaeus vannamei)Cht1 (EU883591.1)、Cht2 (EU861222.1)、Cht3 (AAN74647.1)、Cht4 (FJ888480.1)、Cht5 (FJ888481.1)、Cht6 (GQ916594.1);日本仿长额虾(Pandalopsis japonica) Cht (AFC60660.1)、Cht1 (JF694836.1)、Cht2 (JN982965.1)、Cht3 (JF694838.1)、Cht4 (JF694837.1) |
利用qRT-PCR技术,对三疣梭子蟹PtCht1基因进行了全组织表达分布分析。结果显示,PtCht1在肝胰腺、胃、肠、表皮、肌肉、眼柄、心脏、第6对鳃、血细胞和第1对鳃中均有表达,其中,在肝胰腺中的表达量最高,胃次之,而在其他组织中少有表达(图 5)。
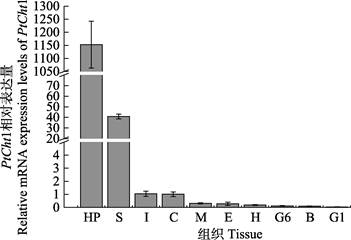
|
图 5 三疣梭子蟹PtCht1基因在不同组织中的表达水平 Fig.5 Relative mRNA expression level of P. trituberculatus PtCht1 gene in different tissues Hp:肝胰腺; S:胃; I:肠; C:表皮; M:肌肉; E:眼柄; H:心脏; G6:第6对鳃; B:血细胞; G1:第1对鳃 Hp: Hepatopancreas; S: Stomach; I: Intestine; C: Cuticle; M: Muscle; E: Eyestalk; H: Heart; G6: Sixth gill; B: Blood cell; G1: First gill |
PtCht1基因在三疣梭子蟹不同蜕皮时期的肝胰腺中的表达模式显示(图 6),PtCht1在蜕皮间期的肝胰腺中的表达量最高,蜕皮后期次之,在蜕皮前期最低,不同蜕皮时期之间表达量差异显著(P < 0.05)。

|
图 6 PtCht1基因在三疣梭子蟹不同蜕皮时期肝胰腺中的表达水平 Fig.6 Relative mRNA expression level of P. trituberculatus PtCht1 gene in hepatopancreas during molting 不同英文字母表示差异显著(P < 0.05).下同 Different letters on the column indicate significant differences (P < 0.05). The same as below |
三疣梭子蟹在进行72 h低盐度(11)胁迫后,PtCht1基因在肝胰腺及第6对鳃中的相对表达量如图 7所示。在肝胰腺中,相比于对照组,胁迫后的PtCht1基因的相对表达量在3 h急剧上升,达到最大值,为对照组的2.72倍,呈显著上调表达(P < 0.05),6 h迅速降低,至24 h均为显著下调表达(P < 0.05),其中,12 h降至最小值,为对照组的0.23倍,48 h再次回升至对照组水平,72 h增至上调表达(P < 0.05)。在第6对鳃组织中,相比对照组,胁迫后的PtCht1基因在3~6 h呈现显著上调表达(P < 0.05),至6 h达到第1个峰值,为对照组的2.67倍,12 h急剧降低至最小值,为对照组的0.46倍,呈显著下调表达(P < 0.05),24 h后逐渐上升,至72 h达到最大值,为对照组的9.96倍,呈显著上调表达(P < 0.05)。
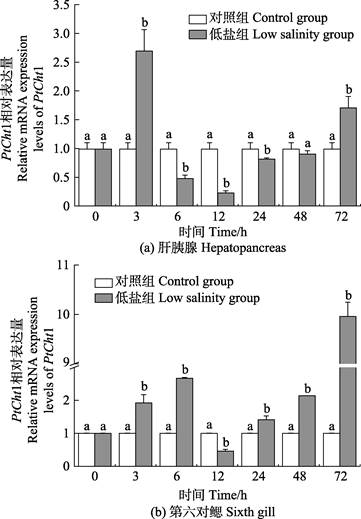
|
图 7 低盐胁迫下三疣梭子蟹PtCht1在肝胰腺和第6对鳃的表达水平 Fig.7 Relative mRNA expression levels of P. trituberculatus PtCht1 gene in the hepatopancreas and sixth gill under low salinity stress |
三疣梭子蟹养殖的不同发育阶段的动物性饵料多以卤虫、杂鱼、蓝蛤等为主,而这一摄食组成中含有大量的几丁质成分,需要几丁质酶进行消化分解后才能吸收(Das et al, 2018; Zhou et al, 2018)。本研究成功克隆三疣梭子蟹几丁质酶基因1 (PtCht1),其编码的蛋白具有含第18家族保守基序的催化结构域、活性位点(FDGLDLDVE)以及几丁质结合结构域ChtBD2 (Boot et al, 2001),且ChtBD2含6个保守的半胱氨酸,可形成3个二硫键,对稳定几丁质酶的结构及调控几丁质酶的活性十分重要(Zhou et al, 2017)。氨基酸比对及系统进化树分析显示,PtCht1与拟穴青蟹SsCht1基因的同源性最高,同时,与其他甲壳动物几丁质酶1紧密聚为一支。由此,根据结构域和活性位点的高度保守性结合聚类分析,确定了PtCht1基因为三疣梭子蟹第18家族几丁质酶基因,且隶属于Group1。
甲壳动物几丁质酶1基因常高表达于肝胰腺中,而肝胰腺是分泌消化酶的主要场所,故几丁质酶1基因在肝胰腺中的功能与几丁质类食物的消化相关(Watanabe et al, 1998)。本研究中,三疣梭子蟹PtCht1基因在肝胰腺和胃等消化器官中的表达量远高于其他组织,表明PtCht1亦可能发挥消化几丁质类食物的功能,这与Zhang等(2010)关于中国对虾的几丁质酶基因FcCht1和FcCht3的报道类似。此外,Zhang等(2014)发现,日本沼虾MnCht1A和MnCht1B基因可能与可溶性几丁质的消化水解有关,因其具有几丁质结合结构域ChtBD2,而PtCht1同样具有该结构,所以PtCht1可能与非可溶性几丁质类食物的消化有关。
斑节对虾几丁质酶PmCht1(Tan, 2000)与日本对虾PjCht1(Watanabe et al, 1996)高度同源,均在肝胰腺中特异性表达,被认为主要与几丁质类食物的降解有关,且在不同的蜕皮阶段,肝胰腺mRNA水平上的表达差异显著。PtCht1在蜕皮间期,表达量显著升高(P < 0.05),但在蜕皮前期和后期有所下降,可能与三疣梭子蟹的生活习性相关。一般情况下,梭子蟹在蜕皮前期和后期阶段摄食停滞,而在蜕皮间期,处于快速生长阶段,摄食活跃,需要更多的几丁质酶消化食物(周凯敏, 2017),进一步表明PtCht1在消化中发挥重要作用。
盐度是影响三疣梭子蟹生长的主要环境因素之一(何鹏等, 2019)。本研究探究了低盐胁迫对几丁质酶基因的表达和三疣梭子蟹生长的影响。结果显示,低盐度会干预PtCht1基因的正常表达,使其在肝胰腺中呈先上调后下调再回升的表达规律,并且PtCht1基因亦能够对盐度的改变做出快速应答,在3 h即达到峰值,而已有研究表明,低盐度条件下,需要机体提供更多的能量来调节渗透压(Ye et al, 2009),由此推断,PtCht1可能在促进食物消化,提供机体所需的能量,进而调控三疣梭子蟹生长及机体应对低盐胁迫过程中具有重要作用(Nikapitiya et al, 2015; Lu et al, 2019)。三疣梭子蟹主要依靠后鳃进行渗透压的调节,PtCht1基因在低盐度胁迫下第6对鳃中的表达具有显著性差异,除12 h外,整体为上调表达,暗示了PtCht1基因可能参与抗低盐胁迫的渗透压调节过程(Lü et al,2013),并发挥积极作用,但详细的调节机制尚需进一步探索。
综上所述,本研究克隆并分析了三疣梭子蟹几丁质酶基因GH18家族Group1成员PtCht1基因的结构特征,丰富了甲壳动物几丁质酶基因家族。通过分析PtCht1基因的组织表达特征,蜕皮周期及低盐度胁迫下的表达模式,探究了基因在三疣梭子蟹消化、生长及渗透压调节方面发挥的生理功能,可为甲壳动物几丁质酶基因的相关调控机理研究提供参考信息,对快速生长及耐低盐新品种的培育提供新途径。
Adrangi S, Faramarzi MA. From bacteria to human:A journey into the world of chitinases. Biotechnology Advances, 2013, 31(8): 1786-1795 DOI:10.1016/j.biotechadv.2013.09.012 |
Boot RG, Blommaart EFC, Swart E, et al. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. Journal of Biological Chemistry, 2001, 276(9): 6770-6778 DOI:10.1074/jbc.M009886200 |
Das S, Vraspir L, Zhou W, et al. Transcriptomic analysis of differentially expressed genes in the molting gland (Y-organ) of the blackback land crab, Gecarcinus lateralis, during molt-cycle stage transitions. Comparative Biochemistry and Physiology Part D Genomics and Proteomics, 2018, 28: 37-53 DOI:10.1016/j.cbd.2018.06.001 |
Duan YF, Liu P, Li JT, et al. Cloning and expression analysis of Cathepsin L cDNA of Exopalaemon carinicauda. Zoological Research, 2013, 34(1): 39-46 [段亚飞, 刘萍, 李吉涛, 等. 脊尾白虾组织蛋白酶L基因的克隆及其表达分析. 动物学研究, 2013, 34(1): 39-46] |
Gao C, Cai X, Zhang Y, et al. Characterization and expression analysis of chitinase genes (CHIT1, CHIT2 and CHIT3) in turbot (Scophthalmus maximus L.) following bacterial challenge. Fish and Shellfish Immunology, 2017, 64: 357-366 DOI:10.1016/j.fsi.2017.03.019 |
Gao L, Sun J, Secundo F, et al. Cloning, characterization and substrate degradation mode of a novel chitinase from Streptomyces albolongus ATCC 27414. Food Chemistry, 2018, 261: 329-336 DOI:10.1016/j.foodchem.2018.04.068 |
Gayathri R, Venkatesh K, Arun M, et al. Bactericidal and fungistatic activity of peptide derived from GH18 domain of prawn chitinase 3 and its immunological functions during biological stress. International Journal of Biological Macromolecules:Structure, Function and Interactions, 2018, 106: 1014-1022 |
He P, Jiang SG, Li YD, et al. Molecular cloning and expression pattern analysis of GLUT1 in black tiger shrimp (Penaeus monodon). South China Fisheries Science, 2019, 15(2): 72-82 [何鹏, 江世贵, 李运东, 等. 斑节对虾GLUT1基因cDNA的克隆与表达分析. 南方水产科学, 2019, 15(2): 72-82] |
Huang QS, Yan JH, Tang JY, et al. Cloning and tissue expressions of seven chitinase family genes in Litopenaeus vannamei. Fish and Shellfish Immunology, 2010, 29(1): 75-81 DOI:10.1016/j.fsi.2010.02.014 |
Li X, Xu Z, Zhou G, et al. Molecular characterization and expression analysis of five chitinases associated with molting in the Chinese mitten crab, Eriocheir sinensis. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 2015, 187: 110-120 DOI:10.1016/j.cbpb.2015.05.007 |
Lü JJ, Liu P, Wang Y, et al. Transcriptome analysis of Portunus trituberculatus in response to salinity stress provides insights into the molecular basis of osmoregulation. PLoS One, 2013, 8(12): e82155 DOI:10.1371/journal.pone.0082155 |
Lu S, Li R, Zheng W, et al. Long-term low-salinity stress affects growth, survival and osmotic related gamma-aminobutyric acid regulation in the swimming crab Portunus trituberculatus. Aquaculture Research, 2019, 50(10): 2888-2895 DOI:10.1111/are.14242 |
Merzendorfer H. Insect chitin synthases:A review. Journal of Comparative Physiology B:Biochemical, Systemic, and Environmental Physiology, 2006, 176(1): 1-15 DOI:10.1007/s00360-005-0005-3 |
Nikapitiya C, Kim WS, Park K, et al. Chitinase gene responses and tissue sensitivity in an intertidal mud crab (Macrophthalmus japonicus) following low or high salinity stress. Cell Stress and Chaperones, 2015, 20(3): 517-526 DOI:10.1007/s12192-015-0576-1 |
Priya TAJ, Li F, Zhang J, et al. Molecular characterization and effect of RNA interference of retinoid X receptor (RXR) on E75 and chitinase gene expression in Chinese shrimp Fenneropenaeus chinensis. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 2009, 153(1): 121-129 DOI:10.1016/j.cbpb.2009.02.009 |
Proespraiwong P, Tassanakajon A, Rimphanitchayakit V. Chitinases from the black tiger shrimp Penaeus monodon:Phylogenetics, expression and activities. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 2010, 156(2): 90-96 |
Qiang Y, Stephen SF. Cloning and characterization of a chitinase from Thermobifida fusca reveals Tfu_0580 as a thermostable and acidic endochitinase. Biotechnology Reports, 2018, 19: e00274 DOI:10.1016/j.btre.2018.e00274 |
Rocha J, Garcia-Carreño FL, Muhlia-Almazán A, et al. Cuticular chitin synthase and chitinase mRNA of whiteleg shrimp Litopenaeus vannamei during the molting cycle. Aquaculture, 2012, 330-333: 111-115 DOI:10.1016/j.aquaculture.2011.12.024 |
Salma U, Uddowla MH, Kim M, et al. Five hepatopancreatic and one epidermal chitinases from a pandalid shrimp (Pandalopsis japonica):Cloning and effects of eyestalk ablation on gene expression. Comparative Biochemistry and Physiology Part B Biochemistry and Molecular Biology, 2012, 161(3): 200-207 |
Shen J, Zhu DF, Hu ZH, et al. Molt staging in the swimming crab Portunus trituberculatus. Journal of Fisheries of China, 2011, 35(10): 1481-1487 [沈洁, 朱冬发, 胡则辉, 等. 三疣梭子蟹蜕皮周期的分期. 水产学报, 2011, 35(10): 1481-1487] |
Song L, Lü JJ, Wang L, et al. Cloning of chitinase gene (PtCht6) in Portunus trituberculatus and its functional analysis in immunity. Oceanologia et Limnologia Sinica, 2019, 50(5): 1080-1090 [宋柳, 吕建建, 王磊, 等. 三疣梭子蟹几丁质酶基因(PtCht6)的克隆及其在免疫中的功能分析. 海洋与湖沼, 2019, 50(5): 1080-1090] |
Sui YM, Gao BQ, Liu P, et al. The tolerance to and optimal salinity for growth in swimming crab Portunus trituberculatus "Huangxuan No.1". Journal of Dalian Ocean University, 2012, 27(5): 398-401 [隋延鸣, 高保全, 刘萍, 等. 三疣梭子蟹"黄选l号"盐度耐受性及适宜生长盐度分析. 大连海洋大学学报, 2012, 27(5): 398-401 DOI:10.3969/j.issn.2095-1388.2012.05.003] |
Tan SH. The Penaeus monodon chitinase 1 gene is differentially expressed in the hepatopancreas during the molt cycle. Marine Biotechnology, 2000, 2(2): 126-135 |
Umemoto N, Ohnuma T, Mizuhara M, et al. Introduction of a tryptophan side chain into subsite+1 enhances transglycosylation activity of a GH-18 chitinase from Arabidopsis thaliana, AtChiC. Glycobiology, 2013, 23(1): 81-90 DOI:10.1093/glycob/cws125 |
Watanabe T, Kono M, Aida K, et al. Isolation of cDNA encoding a putative chitinase precursor in the kuruma prawn Penaeus japonicus. Molecular Marine Biology and Biotechnology, 1996, 5: 299-303 |
Watanabe T, Kono M, Aida K, et al. Purification and molecular cloning of a chitinase expressed in the hepatopancreas of the penaeid prawn Penaeus japonicus.Biochimica et. Biophysica Acta,, 1998, 1382(2): 181-185 DOI:10.1016/S0167-4838(97)00184-2 |
Watanabe T, Kono M. Isolation of a cDNA encoding a chitinase family protein from cuticular tissues of the Kuruma prawn Penaeus japonicus. Zoological Science, 1997, 14(1): 65-68 |
Ye L, Jiang SG, Zhu XM, et al. Effects of salinity on growth and energy budget of juvenile Penaeus monodon. Aquaculture, 2009, 290(1-2): 140-144 DOI:10.1016/j.aquaculture.2009.01.028 |
Zhang F, Lü JJ, Liu P, et al. Cloning and expression analysis of the cDNA of PtCht3 in Portunus trituberculatus. Progress in Fishery Sciences, 2017, 38(2): 167-176 [张凤, 吕建建, 刘萍, 等. 三疣梭子蟹(Portunus trituberculatus)几丁质酶PtCht3基因克隆鉴定及表达分析. 渔业科学进展, 2017, 38(2): 167-176] |
Zhang F, Lü JJ, Liu P, et al. Cloning and expression of chitinase under low salinity stress during molting in Portunus trituberculatus. Oceanologia et Limnologia Sinica, 2015, 46(4): 948-957 [张凤, 吕建建, 刘萍, 等. 三疣梭子蟹几丁质酶基因克隆鉴定及在低盐胁迫和蜕皮周期中的表达分析. 海洋与湖沼, 2015, 46(4): 948-957] |
Zhang J, Kopparapu NK, Yan Q, et al. Purification and characterisation of a novel chitinase from persimmon (Diospyros kaki) with antifungal activity. Food Chemistry, 2013, 138(2-3): 1225-1232 DOI:10.1016/j.foodchem.2012.11.067 |
Zhang J, Sun Y, Li F, et al. Molecular characterization and expression analysis of chitinase (Fcchi-3) from Chinese shrimp, Fenneropenaeus chinensis. Molecular Biology Reports, 2010, 37(4): 1913-1921 |
Zhang SY, Jiang SF, Xiong YW, et al. Six chitinases from oriental river prawn Macrobrachium nipponense:cDNA characterization, classification and mRNA expression during post-embryonic development and moulting cycle. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 2014, 167: 30-40 DOI:10.1016/j.cbpb.2013.09.009 |
Zhou FL, Zhou KM, Huang JH, et al. Characterization and expression analysis of a chitinase gene (PmChi-5) from black tiger shrimp (Penaeus monodon) under pathogens infection and ambient ammonia-N stress. Fish and Shellfish Immunology, 2018, 72: 117-123 DOI:10.1016/j.fsi.2017.10.051 |
Zhou KM, Zhou FL, Huang JH, et al. Characterization and expression analysis of a chitinase gene (PmChi-4) from black tiger shrimp (Penaeus monodon) under pathogen infection and ambient ammonia nitrogen stress. Fish and Shellfish Immunology, 2017, 62: 31-40 DOI:10.1016/j.fsi.2017.01.012 |
Zhou KM. Expression analysis of chitinase genes from Penaeus monodon. Masterxs Thesis of Shanghai Ocean University, 2017 [周凯敏. 斑节对虾几丁质酶基因的克隆与表达. 上海海洋大学硕士研究生学位论文, 2017] |
Zhou ZK, Gu WB, Wang C, et al. Seven transcripts from the chitinase gene family of the mud crab Scylla paramamosain:Their expression profiles during development and moulting and under environmental stresses. Aquaculture Research, 2018, 93: 223-235 |
Zou E, Bonvillain R. Chitinase activity in the epidermis of the fiddler crab, Uca pugilator, as an in vivo screen for molt-interfering xenobiotics. Comparative Biochemistry and Physiology Part C:Toxicology and Pharmacology, 2004, 139(4): 225-230 DOI:10.1016/j.cca.2004.11.003 |



