2. 南方海洋科学与工程广东省实验室(湛江) 广东 湛江 524025
2. Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), Zhanjiang, Guangdong 524025, China
骨的新陈代谢(骨代谢)包括骨的形成和吸收,由成骨、破骨细胞行使功能,主要表现为钙的生物矿化及再活化(钙代谢)。硬骨鱼类的耳石、鳞、鳍及其内骨骼作为鱼体的钙库,涵盖了鱼体约99%的钙(Flik et al, 1986)。鱼类钙的稳态调节主要通过鳃、肠等组织的离子交换及营养吸收作用完成(Loewenac et al, 2016)。当饵料中的钙摄入量不足时,鱼类可通过鳃、肠等组织吸收水环境中的Ca2+进行补充。鱼体对Ca2+的吸收效率还受到盐度的影响。例如,在高盐(如海水)水体中,金头鲷(Sparus aurata)肠道对Ca2+的吸收效率更高(Guerreiro, 2004)。班红琴等(2010)研究发现,虹鳟(Oncorhynchus mykiss)血清Ca2+浓度随盐度的升高呈先降低后升高的变化趋势。由此可见,盐度变化对鱼体内的钙代谢过程具有显著影响。
鳞是鱼类特有的皮肤衍生物,也是鱼类外骨骼的重要组成部分,其细胞组成和骨化机制与内骨骼十分相似。某些硬骨鱼类鳞的钙含量可达鱼体总钙的20%,是鱼类潜在的钙储存库(Persson et al, 1995)。已有研究表明,当鱼体对钙的需求显著增加(如性成熟时期)或钙缺乏较多且不能通过其他途径得到补充时,会通过吸收鳞中的钙(钙活化)进行弥补(Guerreiro et al, 2002)。据报道,金鱼(Carassius auratus)及虹鳟的钙活化优先发生于鳞中,其次为鳍条,而内骨骼为最终选择(Persson et al, 1997; Shinozaki et al, 2000)。由此可见,相较于内骨骼,鳞中的矿化钙更易被调用(Berg, 1968)。此外,鳞作为鱼类的外骨骼,直接暴露于水环境且更易于采集,可作为钙代谢研究的优良载体,还可为进一步揭示盐度变化对鱼类骨代谢的影响提供理想模型。
虹鳟属鲑形目(Salmoniformes)、鲑科(Salmonidae)、太平洋鲑属(大马哈鱼属)(Oncorhynchus),是原产于北美洲的一种冷水性鱼类(贾素文等, 2016)。近年来,全球鲑鳟鱼养殖年产量超过200万t (户国等, 2017),其中,虹鳟是我国养殖产量最高的鲑鳟鱼类。虹鳟属广盐性鱼类,体重在35 g以上的幼鱼经半咸水过渡即可适应海水生活,且海水养殖虹鳟在生长速度、抗病力和肉质等方面更具优势(班红琴等, 2010; 贾素文等, 2016)。我国大部分海域夏季温度过高,不适合虹鳟的网箱养殖,而虹鳟的海水工厂化养殖处于起步阶段,目前仍以淡水养殖为主(杨静雯等, 2019)。因此,虹鳟的盐度驯化是目前虹鳟养殖生产面临的主要问题之一。研究盐度驯化对虹鳟骨代谢的调节机制,不仅可为虹鳟养殖生产实践提供参考,还可为探明硬骨鱼类骨代谢的调控机理及其应对环境变化的演化规律提供新视角。
近年来,针对脊椎动物代谢过程的分子机制研究已成为热点。随着各种组学技术的发展,转录组学(transcriptomics)技术因其在发现基因、查找控制特异性状的潜在主效基因和研究基因表达调控方面最为简单有效,被广泛采用(Li et al, 2014)。本研究利用转录组测序(RNA-Seq)技术筛选与盐度适应及骨代谢相关功能基因,分析盐度驯化对虹鳟鳞组织基因表达水平的影响。通过对差异表达基因(differentially expressed genes, DEGs)进行功能注释和通路富集,获取上述基因的功能分类及显著富集的功能分析结果;采用实时荧光定量PCR(RT-qPCR)技术对盐度驯化下基因表达量进行检测,验证转录组数据的可靠性,相关结果可为鱼类骨代谢调控相关功能基因的挖掘提供基础资料。
1 材料与方法 1.1 实验材料实验用虹鳟幼鱼共计180尾[体长(17.00±1.65) cm,体重(52.90±11.65) g],采集于山东省潍坊市临朐县淡水虹鳟养殖基地。分别将虹鳟幼鱼置于6个有效体积为1000 L (直径1.6 m,高0.6 m)的PE桶中(30尾/桶),水深约为PE桶高度的1/2,流水暂养3 d (淡水,水温为14~16℃,水体硬度约为35.75 mmol/L),光周期条件为12 h: 12 h,每天08:00和16:00投喂配合饲料(总投喂量约为鱼体重的3%)。
本实验设置海水(盐度28)驯化组(seawater group, SW)和常规淡水养殖组(对照,control group, CG) (水体盐度约为3),每组设置3个平行。其中,SW以4/d的盐度提升速率对虹鳟进行盐度驯化,盐度升至28后维持不变。进行盐度驯化的第1天记为1 d,驯化至7 d时进行取样。海水由海水素(青岛海之盐水族科技有限公司)配制。每个平行组分别采集2尾虹鳟鳞组织(所有鳞组织样品均采集于鱼体左侧,背鳍基部下方至尾柄处,面积约1 cm2),立即投入液氮速冻,再转移至–80℃冰箱保存备用。
1.2 总RNA提取及cDNA第一链的合成虹鳟鳞组织,按照Trizol (Invitrogen, 美国)常规法提取总RNA。通过1.5%琼脂糖凝胶电泳和核酸测定仪分别检测总RNA的完整性及浓度,根据PrimeScriptTMⅡ1st strand cDNA synthesis试剂盒(TaKaRa)说明书,将1 μg总RNA反转录合成第一链cDNA,并保存于–40℃冰箱,用于RT-qPCR验证。
1.3 转录组文库构建及高通量测序将质检合格的RNA样品送至杭州联川生物技术股份有限公司完成RNA-Seq文库制备及测序。去除合格样品中核糖体RNA后,以cDNA文库的平均插入尺寸为2×150 bp (PE 150)的规则进行文库构建。将建好的测序文库在Illumina HiSeq 4000平台上按照说明书的要求进行配对测序。
1.4 转录组数据处理与分析将Illumina HiSeq 4000平台上得到的原始图像数据经过Base Calling转化为原始测序序列数据。使用Cutadpter V1.10 (Martin, 2011)对原始数据进行预处理,并使用Fast QC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)在线工具对样本的测序数据质量进行可视化评估。通过一系列的质量控制筛选,得到高质量的clean data,再使用Bowtie 2 (Langmead et al, 2012)和Hisat 2 (Kim et al, 2015)映射读取基因组数据,最后每个样本的映射读本使用StringTie软件(Pertea et al, 2015)进行序列拼接与合并。将经过拼接合并之后的转录本去冗余后,取聚类最长的转录本作为unigene,构建本研究的unigene库。
1.5 差异表达基因筛选及其GO注释和KEGG富集分析采用Bowtie 2 (Langmead et al, 2012)将测序得到的read与unigene库进行比对,基因表达量用FPKM (fragments per kilobase of exon per million fragments mapped)进行估算,计算公式:
| $\;\;\;\;\;\;\;\;\; \rm{FPKM}={\rm{cDNA}\;fragments/mapped\;fragments (millions)}/\\ {\rm{transcript\;lengths}}\;(\rm{kb}) $ |
为了检测基因表达水平,将每个样品中的clean读数映射到组装的转录组以获得每个基因的读数。在筛选过程中,以log2|fold change|≥1作为不同组间DEGs的筛选阈值,P < 0.05具有统计学意义,使用edgeR (Smyth, 2010)进行样本两两之间的差异表达分析,获得2个样品之间的DEGs,来估算各转录本的表达水平。
用Blast2GO软件(默认参数)对得到所有DEGs的GO (gene ontology)注释信息,并对DEGs进行GO功能分类统计。根据GO的3个ontology,对基因的分子功能、所处的细胞位置以及参与的生物过程进行描述(Zhu et al, 2016)。使用Cluster Profiler对DEGs进行KEGG (Kyoto encyclopedia of genes and genomes)通路富集分析,当矫正后的P < 0.05时,表示该功能存在显著富集(Zhu et al, 2016)。
1.6 实时荧光定量PCR分析基于SW和CG虹鳟鳞组织中基因FPKM值变化倍数,在log2|fold change|≥1且P < 0.05的条件下,随机取8个DEGs (表 1),通过RT-qPCR技术检测转录组数据的可靠性。以核糖体18S rRNA基因作为内参,使用Primer Premier 5.0软件,根据基因cDNA序列分别设计相应的特异性引物(表 2),并由生工生物工程(上海)股份有限公司合成,利用LightCycler480 Ⅱ型实时荧光定量PCR仪,根据SYBR® Premix Ex TaqTM (TliRNase H Plus)试剂盒(TaKaRa)说明进行操作,各样品设置3个重复。所有引物均经过扩增效率检测(E > 90%; R2 > 0.990),RT-qPCR产物经测序验证。
|
|
表 1 用于RT-qPCR验证的差异表达基因 Tab.1 Differentially expressed genes for RT-qPCR verification |
|
|
表 2 本实验所用引物 Tab.2 Primers used in this study |
根据测得的Ct值,采用2–ΔΔCt法(邝杰华等, 2020)计算各基因的相对表达量。所得数据均以平均值±标准差(͞x±SD, n=3)表示,使用SPSS 19.0中的单因素方差分析(one-way ANOVA)法及Duncan´s多重比较,分析各基因表达量在2种不同盐度条件下的差异水平,若P < 0.05,表示有显著差异;若P < 0.01,则差异极显著。
2 结果与分析 2.1 转录组测序数据及比对分析对SW (SW-1、SW-2和SW-3)和CG (CG-1、CG-2和CG-3)的虹鳟鳞组织进行RNA-Seq,共获得75.1 Gb clean data。测序样品的数据质量及比对结果详见表 3,各样品的clean data均达到11.22 Gb,Q30碱基百分比均在97.99%以上。将各样品的clean read与虹鳟鱼基因组进行序列比对,比对效率均在86.05%以上。
|
|
表 3 各样本转录组测序数据信息 Tab.3 Information of transcriptomic read of each sample |
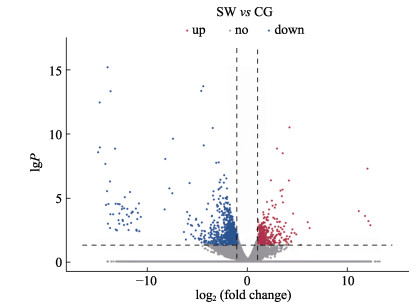
将2组虹鳟鳞组织的测序结果进行比较,在log2|fold change|≥1且P < 0.05条件下,筛出1714个DEGs,其中,上调表达的DEGs数量为484个,下调的数量为1230个,并对DEGs分析结果绘制火山图(图 1)。
|
图 1 差异表达基因的火山图分析 Fig.1 The volcano plot of differentially expressed genes 图中横线为P < 0.05阈值,并且越靠近左边或右边的圆点的表达差异越显著。图中红色圆点表示盐度驯化时上调表达基因,绿色圆点表示盐度驯化时下调表达基因,灰色圆点表示非差异表达基因。 The horizontal line in the figure shows the P < 0.05 threshold, and the closer the dot to the left or the right is, the more significant the difference is. In the figure, the red dot indicates the up-regulated gene, the green dot indicates the down-regulated gene, and the gray dot indicates the non-differentially expressed gene. |
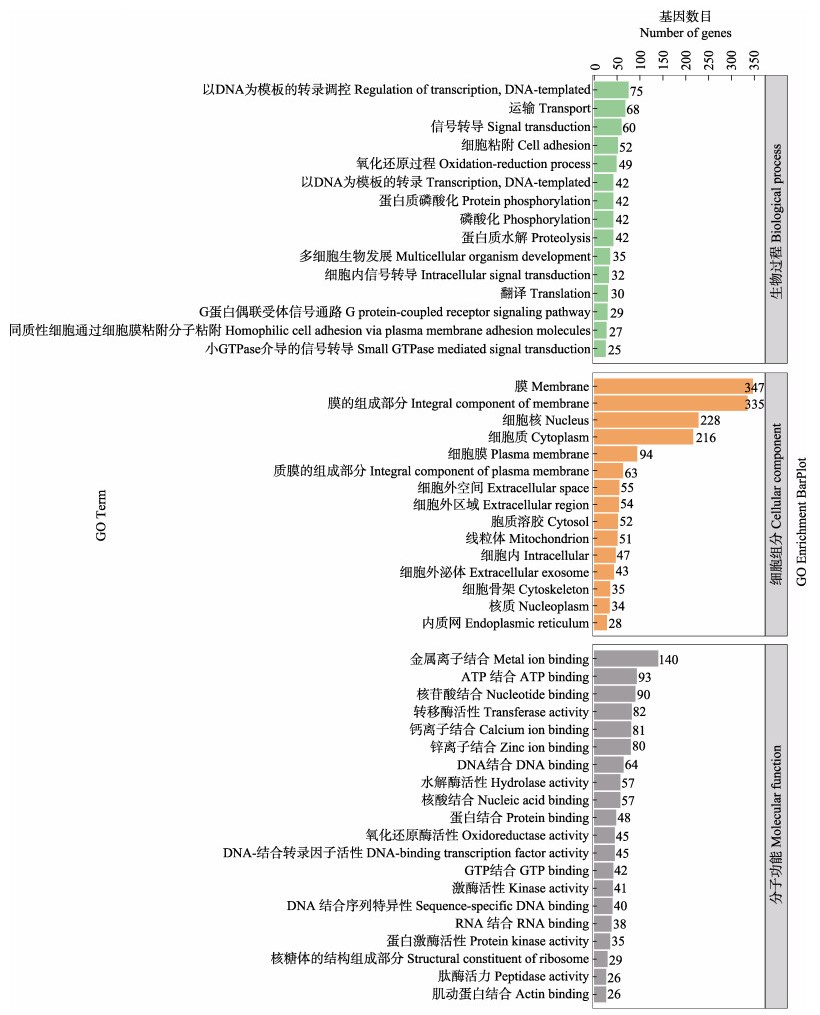
对DEGs进行GO功能注释分析,将所有DEGs归类到GO三大分支:生物过程、细胞组分和分子功能。包括50个功能小类,涉及的生物过程有15个,细胞组分15个,分子功能20个,其中,生物过程部分以DNA为模板的转录调控、运输和信号转导等功能为主;细胞组分中以膜、膜的组成部分、细胞核、细胞质和细胞膜等功能为主;分子功能以金属离子结合、ATP结合、核苷酸结合、钙离子结合和锌离子结合为主(图 2)。
|
图 2 差异表达基因GO富集分析 Fig.2 GO enrichment analysis of differentially expressed genes 纵坐标为富集的GO term,横坐标为该term中差异基因个数。不同颜色用来区分生物过程、细胞组分和分子功能。 The ordinate is the enriched gene ontology (GO) term, and the abscissa is the number of differentially expressed genes in the term. Different colors are used to distinguish biological processes, cellular components and molecular functions. |
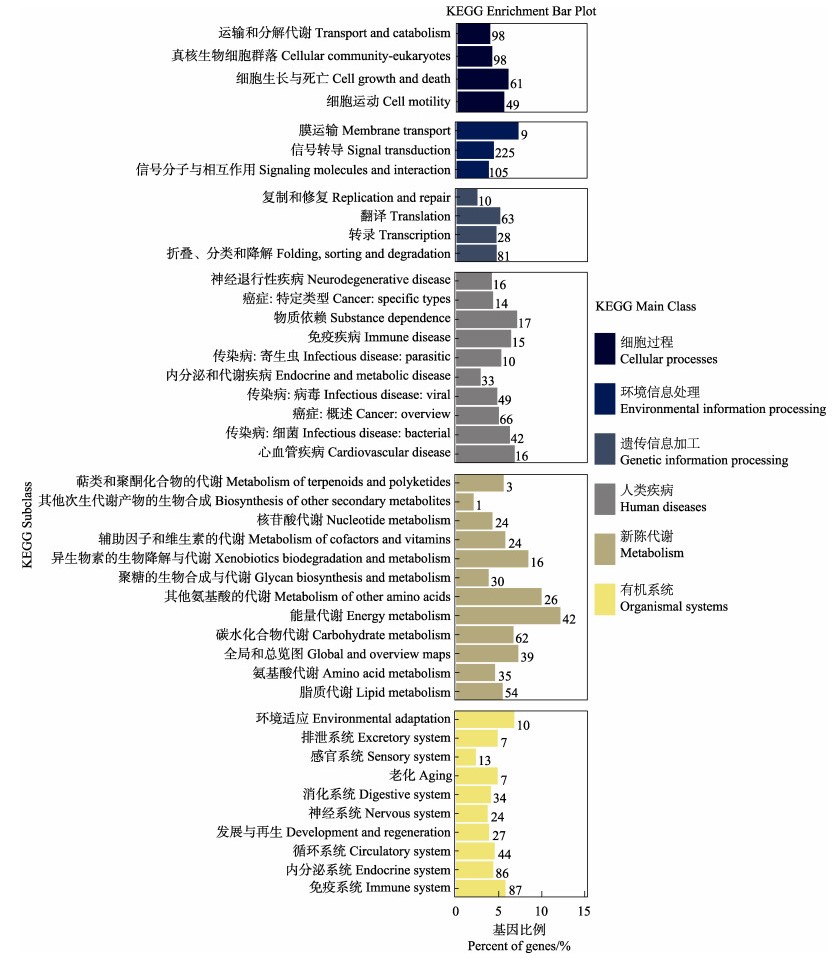
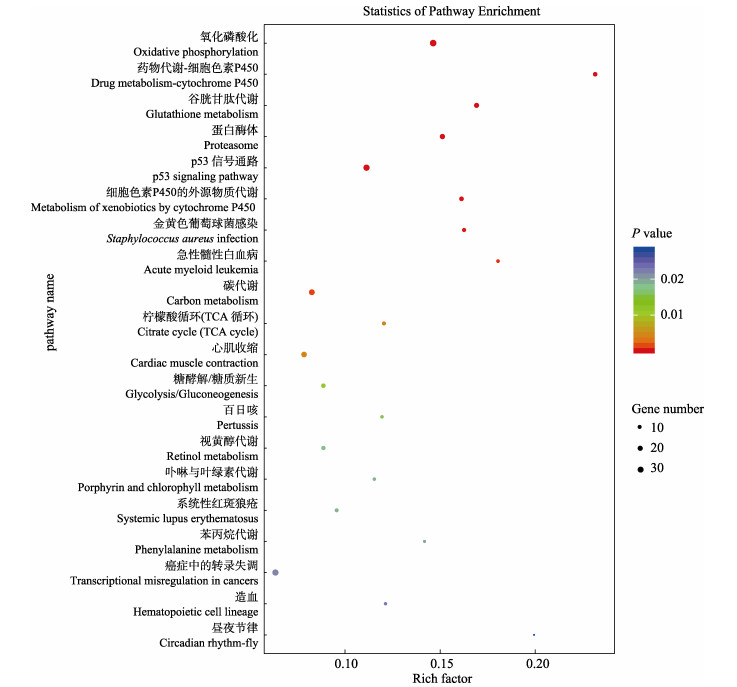
按照KEGG通路类型分类,虹鳟鳞组织DEGs分别注释到细胞过程、环境信息处理、遗传信息加工、人类疾病、新陈代谢和有机系统6个分支(图 3)。其中,虹鳟鳞组织DEGs在新陈代谢、环境信息处理和有机系统这3个一级通路分类(KEGG main class)中富集较多。KEGG二级通路分类(KEGG subclass)富集结果显示,在信号转导、信号分子与相互作用、运输和分解代谢以及真核生物细胞群落等通路中富集的DEGs较多;在能量代谢、其他氨基酸代谢和异生物素生物降解与代谢等通路中富集的DEGs比例较高。在利用KEGG数据库对DEGs进行通路富集分析时,以P < 0.05为阈值,选取了前20条显著富集的代谢通路(图 4)。虹鳟鳞组织的DEGs在氧化磷酸化、药物代谢–细胞色素P450、谷胱甘肽代谢、蛋白酶体、p53信号通路、细胞色素P450对异生物的代谢、金黄色葡萄球菌感染、碳代谢、柠檬酸循环(TCA循环)和心肌收缩等通路中显著富集。其中,参与氧化磷酸化的DEGs数目最多(38个),其次为参与p53信号通路的DEGs (33个)。
|
图 3 差异表达基因KEGG通路富集分析 Fig.3 KEGG pathway enrichment analysis of differentially expressed genes 纵坐标为富集的KEGG二级分类,横坐标为富集到该分类中基因比例,每个二级分类条目上的数字代表该条目上富集到差异基因的数目不同颜色用来区分细胞过程、环境信息处理、遗传信息加工、人类疾病、新陈代谢和有机系统。 The ordinate is the enriched KEGG subclass, and the abscissa is the percent of genes enriched in this subclass, the number on each KEGG subclass item represents the number of differentially expressed genes on that item. Different colors are used to distinguish cellular processes, environmental information processing, genetic information processing, human diseases, metabolism and organismal systems. |
|
图 4 差异表达基因显著富集的前20条KEGG通路 Fig.4 Top 20 KEGG pathways with significant enrichment of differentially expressed genes 圆圈代表一个KEGG通路,纵坐标为通路名称,横坐标为富集因子,圆圈颜色代表P值,颜色越红代表显著富集性越可靠,圆圈越大代表富集的基因数目越多。 Ordinate represents the name of the pathway, and the abscissa represents the enrichment factor, the color of the circle represents P. The deeper the red color, the more reliable the significant enrichment, and the larger the circle, the greater the number of enriched genes. |
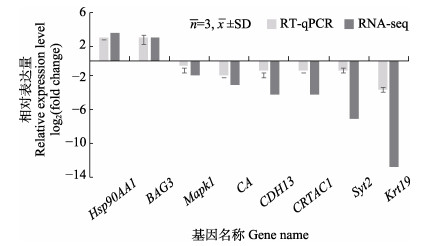
为验证RNA-Seq结果的准确性,本研究采用RT-qPCR对随机挑选的8个DEGs (2个上调和6个下调表达基因)的表达水平进行检测,结果如图 5所示,各基因的表达情况与RNA-Seq结果一致。
|
图 5 差异表达基因的RT-qPCR验证 Fig.5 RT-qPCR verification of differentially expressed genes |
本研究首次对虹鳟鳞组织进行RNA-Seq,结果显示,Q30碱基百分比最低为97.99%,clean read与虹鳟参考基因组序列的比对效率均在86.05%以上。已报道的虹鳟其他组织的RNA-Seq结果中,Q30碱基百分比最低为89.62%,clean read与虹鳟参考基因组序列比对的效率在66.17%以上(李永娟, 2018; Kim, 2021; Cleveland et al, 2021)。相比之下,本研究中虹鳟鳞组织的碱基比对效率较高。
本研究筛选到的DEGs主要为能量代谢、以糖代谢为主的物质代谢过程及渗透压调节相关基因。尽管鳞组织并未直接参与鱼类的渗透压调节过程,但它作为鱼类的皮肤衍生物和外部保护屏障,与水环境接触最充分,在虹鳟盐度驯化的过程中可能发挥了一定作用。由此可见,鳞组织可作为候选对象,用于鱼类环境适应性的调控机制研究。
本研究中,与应激和高渗调节相关的基因,如热休克蛋白90a (heat shock protein HSP 90-alpha, Hsp90a)和BAG家族分子伴侣调节剂3 (BAG family molecular chaperone regulator 3, BAG3)等基因的表达水平显著上调。Hsp90是细胞应激和内源性保护蛋白的生物标志物(Cqza et al, 2021),其表达情况反应了鱼类的应激程度。据报道,盐度胁迫处理下的三疣梭子蟹(Portunus trituberculatus) (Zhang et al, 2009)、大西洋鲑(Salmo salar) (Pan et al, 2000)和大黄鱼(Pseudosciaena crocea) (李明云等, 2015)等物种,其HSP90a mRNA的表达量均显著增加。此外,将三疣梭子蟹HSP90a蛋白转入大肠杆菌(Escherichia coli)可显著增强大肠杆菌的盐度耐受性(覃烨等, 2012)。本研究中,推测虹鳟通过上调Hsp90a提高其盐度适应性。BAG3作为一种存活蛋白,通常在细胞受到如高温、高盐、重金属和蛋白酶体抑制等刺激时被激活。推测在鱼类适应盐度变化的过程中,BAG3的上调可能通过促进蛋白质的识别和转运,提高机体损伤或变性蛋白质代谢水平(Minoia et al, 2014)。此外,与低渗调节相关的碳酸酐酶(carbonic anhydrase, CA)基因显著下调。CA是一类含锌金属的酶,具有促进鱼体对Na+吸收的作用(冉凤霞等, 2020)。斑马鱼(Danio rerio) CA2a和CA15a在低Na+水体中表达显著下调,在高Na+水体中表达显著上调,表明其在鱼类应对低渗环境过程中可能发挥重要作用(Ito et al, 2013)。
虹鳟在从淡水驯化(过渡)至海水(盐度28)的过程中,与Ca2+结合和运输相关基因,如钙粘着蛋白13 (Cadherin 13, CDH13)、软骨酸性蛋白2 (cartilage acidic protein 1, CRTAC1)和突触结合蛋白2 (Synaptotagmin 2, Syt2)等基因的表达被抑制。推测其原因,可能是高渗环境导致鱼体Ca2+的吸收低于排出速率,Ca2+处于较低水平,机体为维持钙稳态的平衡,从而抑制了钙代谢相关基因的表达(班红琴等, 2010)。
已有研究表明,Mmp-2、Mmp-9、Acp5b、Ctsk、TNF-α、Alpl、Osterix、Osteocalcin、OPG和Col12a1等(Kitaura et al, 2020; Metz et al, 2014; Vrieze et al, 2011)基因均有可能参与骨形成、骨吸收及成骨细胞和破骨细胞活性的调节,但RNA-Seq结果显示,这些基因在虹鳟鳞组织中的表达水平未见显著变化。据此推测,本研究所采用的盐度驯化方式未对骨代谢相关基因的表达水平产生显著影响,可见虹鳟作为广盐性鱼类,对水体盐度变化的适应能力较强,在较为缓慢的盐度驯化模式下其骨代谢水平较为稳定。
虹鳟鳞组织DEGs的KEGG通路富集结果显示,DEGs在蛋白酶体通路和能量代谢相关通路上被显著富集。蛋白酶体的主要作用是降解细胞内受到损伤的蛋白质(Schubert et al, 2020)。盐度变化会影响鱼类的渗透压调节系统,而由此产生的细胞内液离子浓度的改变则可能会导致某些蛋白质损伤甚至变性,这种情况下,鱼体可能通过调节蛋白酶体通路相关基因的表达量进行响应(孟玮等, 2021)。如具有蛋白质转运功能的BAG3 mRNA表达水平增强,可进一步提高变性或损伤蛋白质的降解效率。孟玮等(2021)对日本黄姑鱼(Nibea japonica)进行急性盐度胁迫后,筛选到的DEGs在急性炎症反应条目显著富集,表明鱼体在胁迫下产生强烈应激反应。尽管本研究中部分DEGs在药物代谢–细胞色素P450、p53信号通路和急性髓性白血病等与机体免疫和损伤相关的通路上也得到显著富集,但相关基因数量较少,且这些基因并未富集到较多的应激相关通路。这些结果从另一角度证明了本研究选取的驯化模式较为安全合理。
最后,虹鳟鳞组织中的unigene多被富集在MAPK、Wnt和钙信号通路等与骨代谢相关通路中,表明鳞组织可作为骨代谢研究中外骨骼部分的理想模型。然而,DEGs在NF-kB、MAPK (JNK、p38、ERK1/2和STAT3)、Wnt/β-catenin、BMP/Smads、Notch、Jak/Sart、TGF-β和OPG-RANK-RANKL等骨代谢相关信号通路富集不显著(Boyle et al, 2003; Kitaura et al, 2020; 熊燕琴等, 2014),表明以4/d的盐度提升速率对虹鳟进行盐度驯化,未对其鳞组织的骨代谢相关基因表达水平产生显著影响。
BAN H Q, WU Y, LI Y, et al. The changes in osmotic pressure, concentrations of hormones, and ion composition in serum of rainbow trout during salinity acclimation. Journal of Dalian Ocean University, 2010, 25(6): 551-555 [班红琴, 吴垠, 李阳, 等. 盐度驯化过程中虹鳟血清渗透压、激素水平及离子组成的变化. 大连海洋大学学报, 2010, 25(6): 551-555 DOI:10.3969/j.issn.1000-9957.2010.06.014] |
BERG A. Studies on the metabolism of calcium and strontium in freshwater fish. I. Relative contribution of direct and intestinal absorption. Memorie dell'Istituto Italiano di Idrobiologia, 1968, 23: 161-196 |
BOYLE W J, SIMONET W S, LACEY D L. Osteoclast differentiation and activation. Nature, 2003, 423(6937): 337-342 DOI:10.1038/nature01658 |
CLEVELAND B M, GAO G, RADLER L M, et al. Hepatic fatty acid and transcriptome profiles during the transition from vegetable-to fish oil-based diets in rainbow trout (Oncorhynchus mykiss). Lipids, 2021, 56: 189-200 DOI:10.1002/lipd.12287 |
CQZA D, WEI K B, WKY C, et al. The effect of acute heat stress on the innate immune function of rainbow trout based on the transcriptome. Journal of Thermal Biology, 2021, 96(1): 102834 |
FLIK G, FENWICK J C, KOLAR Z, et al. Effects of low ambient calcium levels on whole-body Ca2+ flux rates and internal calcium pools in the freshwater cichlid teleost, Oreochromis mossambicus. Journal of Experimental Biology, 1986, 120(1): 249-264 DOI:10.1242/jeb.120.1.249 |
GUERREIRO P M, FUENTES J M, CANARIO A, et al. Calcium balance in sea bream (Sparus aurata): The effect of oestradiol-17beta. Journal of Endocrinology, 2002, 173(2): 377-385 DOI:10.1677/joe.0.1730377 |
GUERREIRO P M. Water calcium concentration modifies whole-body calcium uptake in sea bream larvae during short-term adaptation to altered salinities. Journal of Experimental Biology, 2004, 207(4): 645-653 DOI:10.1242/jeb.00765 |
HU G, WANG B Q. An overview of global rainbow trout breeding industry with insight into reference to China. Chinese Journal of Fisheries, 2017, 30(3): 1-6 [户国, 王炳谦. 国际虹鳟育种产业简介及其对我国的借鉴意义. 水产学杂志, 2017, 30(3): 1-6 DOI:10.3969/j.issn.1005-3832.2017.03.001] |
ITO Y, KOBAYASHI S, NAKAMURA N, et al. Close association of carbonic anhydrase (CA2a and CA15a), Na+/H+ exchanger (Nhe3b), and ammonia transporter rhcg1 in Zebrafish ionocytes responsible for Na+ uptake. Frontiers in Physiology, 2013, 4: 59 |
JIA S W, LIU L. Adaptability of rainbow trout fry of different sizes to changes in salinity gradient. Hebei Fisheries, 2016(3): 11, 41 [贾素文, 刘丽. 不同规格虹鳟鱼苗对盐度梯度变化适应能力. 河北渔业, 2016(3): 11, 41] |
KIM D, LANGMEAD B, SALZBERG S L. HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 2015, 12(4): 357-360 DOI:10.1038/nmeth.3317 |
KIM J. Transcriptome profiling in head kidney of rainbow trout (Oncorhynchus mykiss) after infection with the low-virulent Nagano genotype of infectious hematopoietic necrosis virus. Archives of Virology, 2021, 166(4): 1057-1070 DOI:10.1007/s00705-021-04980-9 |
KITAURA H, MARAHLEH A, OHORI F, et al. Osteocyte- related cytokines regulate osteoclast formation and bone resorption. International Journal of Molecular Sciences, 2020, 21(14): 5169 DOI:10.3390/ijms21145169 |
KUANG J H, MA Q, MAO F F, et al. Effects of low salinity stress on the expression profiling of HSPB1, HSPB7, and HSPB11 in the roughskin sculpin (Trachidermus fasciatus). Journal of Fishery Sciences of China, 2020, 27(5): 494-503 [邝杰华, 马骞, 毛非凡, 等. 低盐胁迫下松江鲈HSPB1、HSPB7和HSPB11基因的表达变化规律. 中国水产科学, 2020, 27(5): 494-503] |
LANGMEAD B, SALZBERG S L. Fast gapped-read alignment with Bowtie 2. Nature Methods, 2012, 9(4): 357-359 DOI:10.1038/nmeth.1923 |
LI E C, LI C. Use of RNA-seq in aquaculture research. Poultry Fisheries and Wildlife Sciences, 2014, 2(2): 1000e108 |
LI Y J. Transcriptome analysis of liver and head kidney in rainbow trout (Oncorhynchus mykiss) reveals the response to heat stress by RNA-seq. Gansu Agricultural University, 2018 [李永娟. 基于RNA-seq的热应激条件下虹鳟肝脏和头肾转录组分析. 甘肃农业大学, 2018]
|
LI Y M, MIAO L, ZHANG H, et al. Effects of low salt stress on expression of gh, igf-1, hsp90 and pparβ gene in Pseudosciaena crocea. Journal of Ningbo University (Natural Science and Engineering), 2015, 28(4): 1-6 [李明云, 苗亮, 张浩, 等. 低盐协迫对大黄鱼gh、igf-1、hsp90和pparβ基因表达变化的影响. 宁波大学学报(理工版), 2015, 28(4): 1-6] |
LOEWENAC T N, CARRIEREB B, REISTC J D, et al. Review: Linking physiology and biomineralization processes to ecological inferences on the life history of fishes. Comparative Biochemistry and Physiology Part A, Molecular and Integrative Physiology, 2016, 202: 123-140 DOI:10.1016/j.cbpa.2016.06.017 |
MARTIN M. Cutadapt removes adapter sequences from high- throughput sequencing reads. Embnet Journal, 2011, 17(1): 10-12 DOI:10.14806/ej.17.1.200 |
MENG W, XU K D, LI Z H, et al. Transcriptome analysis of Nibea japonica under acute salinity stress. Journal of Fisheries of China, 2021, 45(5): 649-660 [孟玮, 徐开达, 李振华, 等. 急性盐度胁迫对日本黄姑鱼肌肉组织转录组的影响. 水产学报, 2021, 45(5): 649-660] |
METZ J R, LEEUWIS R, ZETHOF J, et al. Zebrafish (Danio rerio) in calcium-poor water mobilise calcium and phosphorus from scales. Journal of Applied Ichthyology, 2014, 30(4): 671-677 DOI:10.1111/jai.12513 |
MINOIA M, BONCORAGLIO A, VINET J, et al. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy, 2014, 10(9): 1603-1621 DOI:10.4161/auto.29409 |
PAN F, ZARATE J M, TREMBLAY G C, et al. Cloning and characterization of salmon hsp90 cDNA upregulation by thermal and hyperosmotic stress. Journal of Experimental Zoology, 2000, 287(3): 199-212 DOI:10.1002/1097-010X(20000801)287:3<199::AID-JEZ2>3.0.CO;2-3 |
PERSSON P, TAKAGI Y, BJÖRNSSON B T. Tartrate resistant acid phosphatase as a marker for scale resorption in rainbow trout, Oncorhynchus mykiss: Effects of estradiol-17β treatment and refeeding. Fish Physiology and Biochemistry, 1995, 14(4): 329-339 DOI:10.1007/BF00004071 |
PERSSON P, JOHANNSSON S H, TAKAGI Y, et al. Estradiol-17β and nutritional status affect calcium balance, scale and bone resorption, and bone formation in rainbow trout, Oncorhynchus mykiss. Journal of Comparative Physiology B Biochemical Systems and Environmental Physiology, 1997, 167(7): 468-473 DOI:10.1007/s003600050098 |
PERTEA M, PERTEA G M, ANTONESCU C M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology, 2015, 33(3): 290-295 DOI:10.1038/nbt.3122 |
QIN Y, XU Q H. The prokaryotic expression of recombinant heat shock protein HSP90a of Portunus trituberculatus under salinity stress. Journal of Fisheries of China, 2012, 36(5): 681-685 [覃烨, 许强华. 盐度胁迫下三疣梭子蟹热休克蛋白HSP90a的原核表达. 水产学报, 2012, 36(5): 681-685] |
RAN F X, JIN W J, HUANG S, et al. Research progress on the effects of salinity change on fish. Journal of Northwest A & F University (Natural Science), 2020, 48(8): 10-18 [冉凤霞, 金文杰, 黄屾, 等. 盐度变化对鱼类影响的研究进展. 西北农林科技大学学报(自然科学版), 2020, 48(8): 10-18] |
SHINOZAKI F, MUGIYA Y. Effects of salmon calcitonin on calcium deposition on and release from calcified tissues in fed and starved goldfish Carassius auratus. Fisheries Science, 2000, 66(4): 695-700 |
SMYTH G K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010, 26(1): 139-140 |
SCHUBERT U, ANTÓN L C, GIBBS J, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature, 2000, 404(6779): 770-774 |
VRIEZE E D, SHARIF F, METZ J R, et al. Matrix metalloproteinases in osteoclasts of ontogenetic and regenerating zebrafish scales. Bone, 2011, 48(4): 704-712 |
XIONG Y Q, ZHOU J, LEI T. Research progress in the signal pathways in bone metabolism. Chinese Journal of Osteoporosis, 2014, 20(2): 200-204 [熊燕琴, 周筠, 雷涛. 骨代谢信号通路的研究进展. 中国骨质疏松杂志, 2014, 20(2): 200-204] |
YANG J W, ZHOU Y G, HUANG M, et al. Comparative studies on digestive and antioxidant enzyme activities between juvenile rainbow (Oncorhynchus mykiss) and steelhead trout (O. mykiss). Periodical of Ocean University of China (Natural Sciences), 2019, 49(3): 122-131 [杨静雯, 周演根, 黄铭, 等. 盐度对虹鳟和硬头鳟幼鱼消化酶和抗氧化酶活性的比较研究. 中国海洋大学学报(自然科学版), 2019, 49(3): 122-131] |
ZHANG X Y, ZHANG M Z, ZHENG C J, et al. Identification of two hsp90 genes from the marine crab, Portunus trituberculatus and their specific expression profiles under different environmental conditions. Comparative Biochemistry and Physiology Part C: Toxicology and Pharmacology, 2009, 150(4): 465-473 |
ZHU W B, WANG L M, DONG Z J, et al. Comparative transcriptome analysis identifies candidate genes related to skin color differentiation in red tilapia. Scientific Reports, 2016, 6(1): 31347 |



