属鱼类物种鉴定中的应用
属鱼类物种鉴定中的应用
2. 中国水产科学研究院 黄海水产研究所 青岛海洋科学与技术试点国家实验室深蓝渔业工程联合实验室 山东 青岛 266071;
3. 海阳市黄海水产有限公司 山东 烟台 265122
 属(Seriola)鱼类快速有效的分子鉴别技术,从DNA条形码角度出发,分析了线粒体细胞色素b (Cyt b)、NADH脱氢酶(ND1和ND2)基因在黄条
属(Seriola)鱼类快速有效的分子鉴别技术,从DNA条形码角度出发,分析了线粒体细胞色素b (Cyt b)、NADH脱氢酶(ND1和ND2)基因在黄条 (Seriola lalandi)、高体
(Seriola lalandi)、高体 (Seriola dumerili)和五条
(Seriola dumerili)和五条 (Seriola quinqueradiata)等
(Seriola quinqueradiata)等 属鱼类物种鉴定和系统进化以及地理区域鉴别中的适用性。结果显示,Cyt b基因表现出明显的A+T偏倚性,ND2基因序列突变速率较高,变异率为20.52%,ND2基因(Hd=0.900, Pi=0.082)的遗传多样性高于ND1 (Hd=0.874, Pi=0.077)和Cyt b (Hd=0.814,Pi=0.061)。比较了
属鱼类物种鉴定和系统进化以及地理区域鉴别中的适用性。结果显示,Cyt b基因表现出明显的A+T偏倚性,ND2基因序列突变速率较高,变异率为20.52%,ND2基因(Hd=0.900, Pi=0.082)的遗传多样性高于ND1 (Hd=0.874, Pi=0.077)和Cyt b (Hd=0.814,Pi=0.061)。比较了 属鱼类3种基因序列的结构特征,基于ND1和ND2基因计算的
属鱼类3种基因序列的结构特征,基于ND1和ND2基因计算的 属鱼类种间遗传距离都为种内遗传距离的10倍以上,但Cyt b基因对高体
属鱼类种间遗传距离都为种内遗传距离的10倍以上,但Cyt b基因对高体 和几内亚
和几内亚 (Seriola carpenteri)辨识力不足。系统进化分析显示,每个物种都形成单系分支,3个基因均能对我国3种
(Seriola carpenteri)辨识力不足。系统进化分析显示,每个物种都形成单系分支,3个基因均能对我国3种 属鱼类进行鉴别,且都可有效区别来自全球3个不同水域的黄条
属鱼类进行鉴别,且都可有效区别来自全球3个不同水域的黄条 种群。因此,Cyt b、ND1和ND2基因不仅可作为
种群。因此,Cyt b、ND1和ND2基因不仅可作为 属鱼类物种鉴定的有效DNA条形码,还可作为不同地理种群划分和种质资源科学保护的依据,为我国
属鱼类物种鉴定的有效DNA条形码,还可作为不同地理种群划分和种质资源科学保护的依据,为我国 鱼养殖产业的持续健康发展和种质资源的可持续利用提供技术依据。
鱼养殖产业的持续健康发展和种质资源的可持续利用提供技术依据。 属鱼类 DNA条形码 物种鉴定 系统进化分析
属鱼类 DNA条形码 物种鉴定 系统进化分析 2. Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Joint Laboratory for Deep Blue Fishery Engineering of Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, Shandong 266071, China;
3. Haiyang Yellow Sea Fishery Company, Yantai, Shandong 265122, China










线粒体DNA具有长度短、结构简单、排列紧凑、偏母性遗传和进化速度快的特性,目前,作为理想的分子标记被广泛应用在鱼类分子系统学、生态地理学和种群遗传学等领域(Wilson et al, 2010; 袁娟等, 2008)。DNA条形码作为分子鉴定中最常见、最直接、最准确的方法,是指生物体内能够代表该物种的、标准的、有足够变异的、易扩增且相对较短的DNA片段,最早由Barrent等(2005)提出,在物种鉴定和进化分析上都有较好的应用(Sepúlveda et al, 2019),弥补了传统形态学鉴定方法的不足,其中,以线粒体细胞色素氧化酶亚基Ⅰ(CO Ⅰ)应用于鱼类物种鉴定最为广泛(王敏等, 2015)。随着条形码技术的不断发展,进化速度适中的Cyt b和NADH脱氢酶基因(ND基因),也被用于亲缘关系较近的种间和种内的遗传差异分析(Clayton, 1992; 陈四海等, 2011)。Cyt b在不同物种中表现出明显的遗传差异。毕潇潇等(2009)通过线粒体16S rRNA、CO I和Cyt b基因片段比较分析,为4种鳕鱼(Gadidae)的鉴定提供了鉴别依据。ND基因在鱼类不同分类阶元的系统发育研究中发挥作用(Serb et al, 2003; Bowen et al, 2008)。梁日深等(2014)通过NADH脱氢酶亚基Ⅰ(ND1)基因分析了16种胡椒鲷属(Plectorhinchus)鱼类的亲缘关系;高志远(2013)分析了我国17个地理群体乌鳢(Channa argus)线粒体NADH脱氢酶亚基Ⅱ(ND2)序列的遗传多样性,为乌鳢分类和系统进化提供了依据。本研究探讨线粒体Cyt b、ND1和ND2基因在

我国3种





取胸鳍组织样品25 mg,用解剖剪剪碎置于无菌离心管中,采用天根海洋动物组织DNA试剂盒提取DNA,参照说明书进行操作。通过琼脂糖凝胶电泳和核酸蛋白测定仪测定DNA浓度和纯度,–20℃保存。
1.2.2 PCR扩增根据序列比对分别设计Cyt b、ND1和ND2基因扩增特异引物(表 1)。3个基因的PCR扩增体系均为50 μL,包括rTaq酶25 μL、模板2 μL、上下游引物各1 μL和ddH2O 21 μL,PCR扩增条件程序见表 2。产物经检测合格后送生工生物工程(上海)股份有限公司双向测序。
|
|
表 1 3种 |
|
|
表 2 3种基因PCR扩增条件 Tab.2 PCR amplification conditions of three genes |
将获得的测序结果采用DNAMAN和DNAstar软件进行序列拼接和校正,并在NCBI数据库进行BLAST比对。从GenBank数据库下载与本研究3种基因扩增区段一致的鲹科鱼类Cyt b、ND1和ND2基因序列,采用DnaSP5.10软件进行遗传多样性参数分析,包括单倍型数(h)、单倍型多样性指数(Hd)、核苷酸多样性指数(Pi)、平均核苷酸差异数(k)等。运用MEGA 7.0软件,统计序列碱基组成、密码子位点偏好性,计算序列保守位点、简约信息位点和变异位点等,运用最大似然法(maximum- likelihood, ML),自举检验1000次,采用Kimura 2-parameter模型计算遗传距离,并构建系统发育树。
2 结果与分析 2.1 基因序列分析3种



|
|
表 3 3种 |
3个基因密码子碱基含量见表 4。Cyt b序列中,第3密码子位点GC含量(56.6%)显著高于第1和第2密码子位点(46.8%和39.4%),且第1密码子变异位点最高(33)。ND1序列中,第3密码子位点GC含量(43.9%)低于第1和第2密码子位点(52.7%和55.0%)。ND2序列中,第3密码子位点GC含量(44.6%)低于第1和第2密码子位点(51.7%和54.0%)。ND1各密码子变异位点相差不大,ND2第1密码子变异位点最低(54),碱基使用频率Cyt b基因表现出明显A+T偏倚性。
|
|
表 4 3种 |
3种
|
|
表 5 3种基因片段的遗传多样性参数 Tab.5 Genetic diversity parameters of three gene fragments |
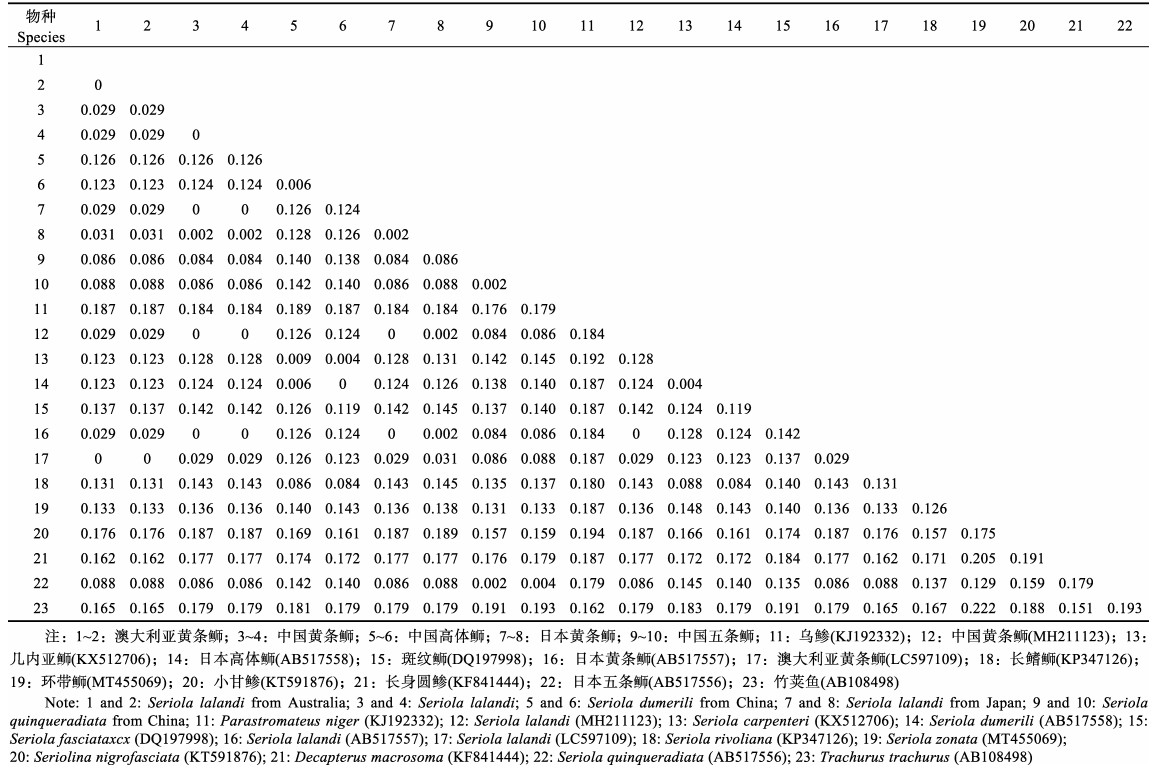
基于Cyt b基因的







|
|
表 6 基于Cyt b的遗传距离分析 Tab.6 Genetic distance analysis based on Cyt b |
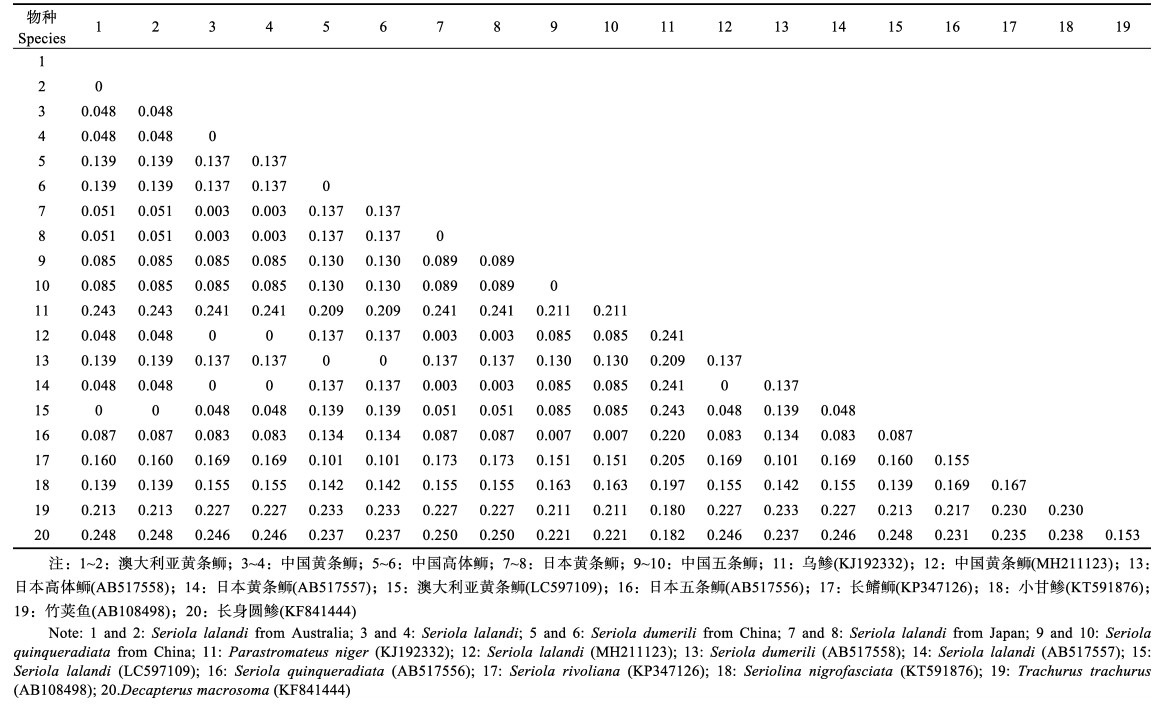
基于ND1基因的







|
|
表 7 基于ND1的遗传距离分析 Tab.7 Genetic distance analysis based on ND1 |
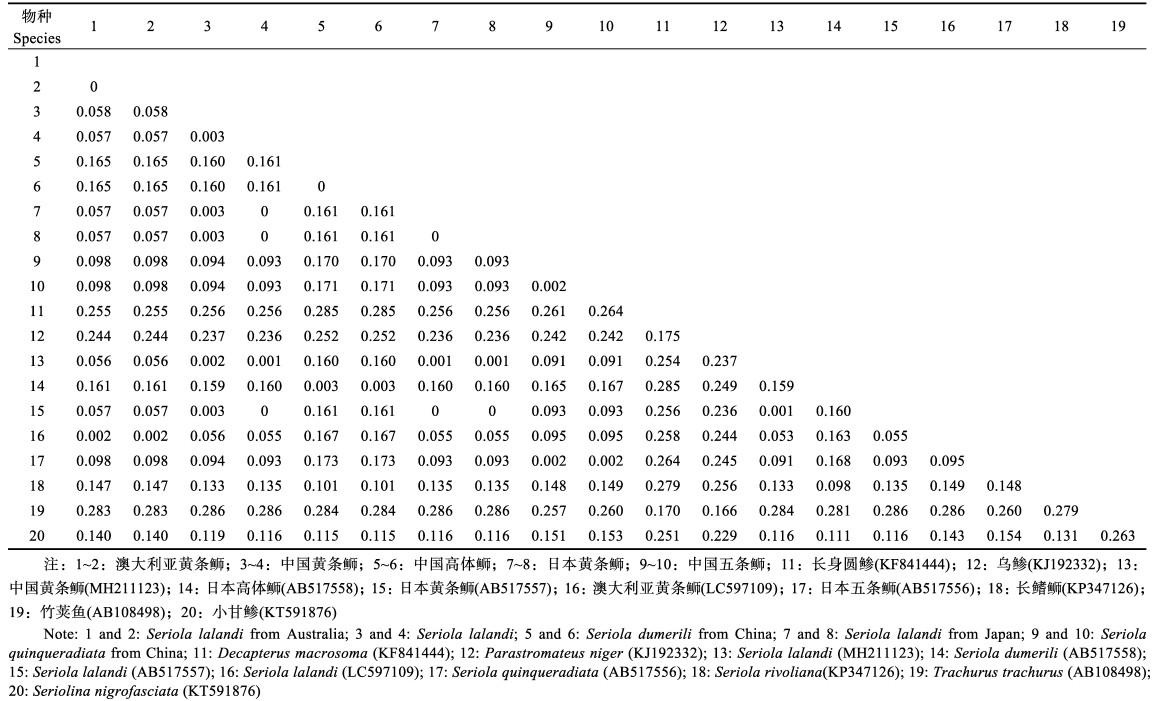
基于ND2基因的








|
|
表 8 基于ND2的遗传距离分析 Tab.8 Genetic distance analysis based on ND2 |
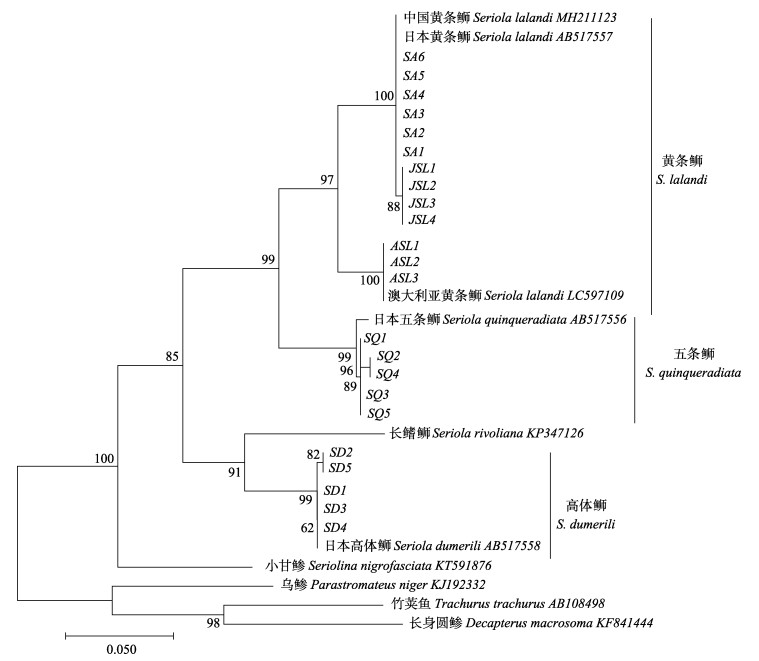
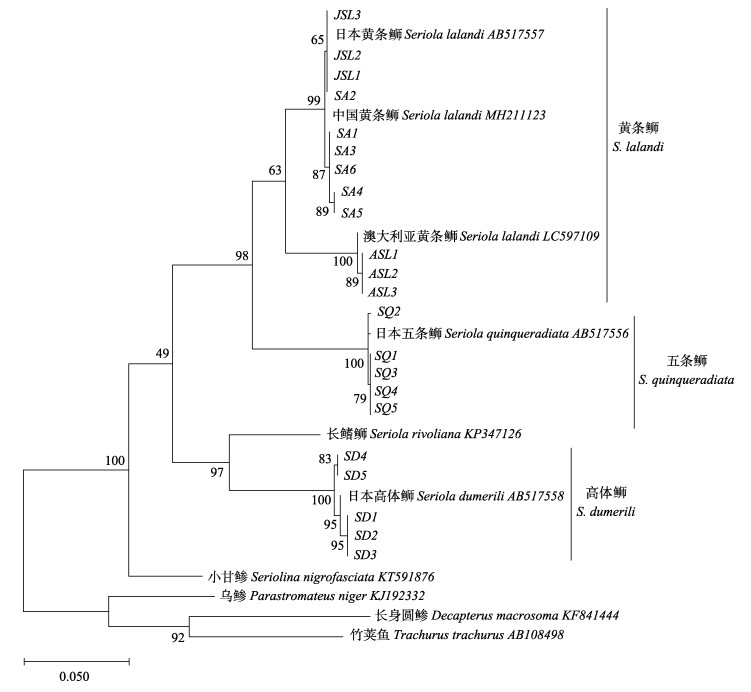
由基于Cyt b、ND1和ND2基因序列构建的










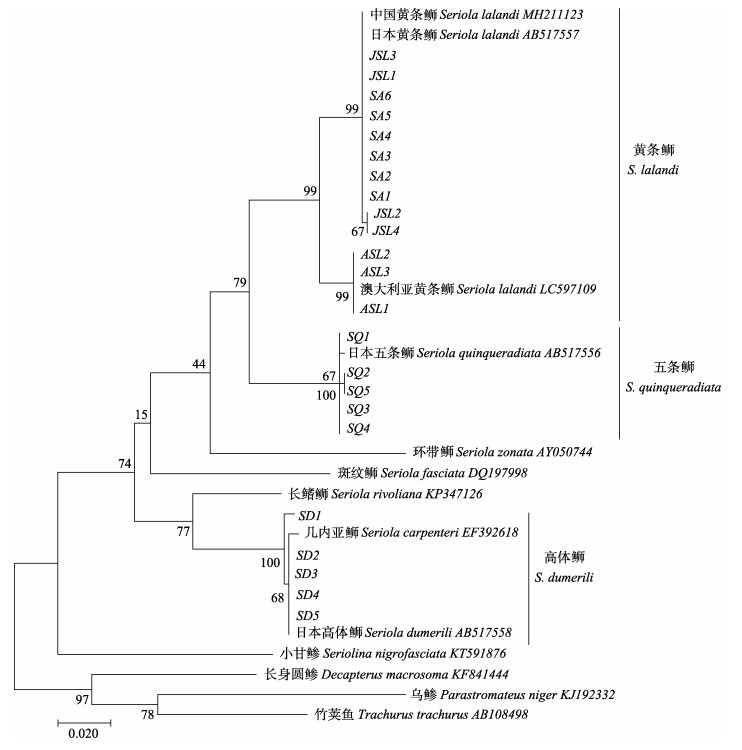
|
图 1 基于Cyt b基因构建的ML系统进化树(枝上数字为置信度值)
Fig.1 Maximum-likelihood phylogenetic tree based on Cyt b gene (The number on branch is confidence value)
SL:中国黄条 ;JSL:日本黄条 ;JSL:日本黄条 ;ASL:澳大利亚黄条 ;ASL:澳大利亚黄条 ;SQ:中国五条 ;SQ:中国五条 ;SD:中国高体 ;SD:中国高体 SL: Seriola lalandi from China; JSL: Seriola lalandi from Japan; ASL: Seriola lalandi from Australia; SQ: Seriola quinqueradiata from China; SD: Seriola dumerili from China
SL: Seriola lalandi from China; JSL: Seriola lalandi from Japan; ASL: Seriola lalandi from Australia; SQ: Seriola quinqueradiata from China; SD: Seriola dumerili from China
|
DNA分子本身含有丰富的多态性信息,受外界环境和其他因素的影响较小,能够准确和稳定地反映物种间的遗传关系,加之线粒体DNA自身所具有的优点,在进行鱼类群体遗传研究中常被作为一种非常重要的分子标记(乔慧莹, 2014)。线粒体DNA序列的进化变异主要包括碱基的转换、倒置和易位等,可在任意时期进行检测,不需要考虑表达的影响因素,检测方法简单、高效(Sanger et al, 2003)。线粒体基因组不同编码基因的进化速率是不同的,各基因所表现出来的遗传信息量也不同(赵凯, 2006)。本研究共分析了Cyt b、ND1与ND2三个线粒体基因在







|
图 2 基于ND1基因构建的ML系统进化树(枝上数字为置信度值)
Fig.2 Maximum-likelihood phylogenetic tree based on ND1 gene (The number on branch is confidence value)
SL:中国黄条 ;JSL:日本黄条 ;JSL:日本黄条 ;ASL:澳大利亚黄条 ;ASL:澳大利亚黄条 ;SQ:中国五条 ;SQ:中国五条 ;SD:中国高体 ;SD:中国高体 SL: Seriola lalandi from China; JSL: Seriola lalandi from Japan; ASL: Seriola lalandi from Australia; SQ: Seriola quinqueradiata from China; SD: Seriola dumerili from China
SL: Seriola lalandi from China; JSL: Seriola lalandi from Japan; ASL: Seriola lalandi from Australia; SQ: Seriola quinqueradiata from China; SD: Seriola dumerili from China
|
ND1和ND2基因进化速率较快,能较好地反映出种内与群体间的遗传变化差异及系统分类地位,已被广泛用于节肢动物、鱼类、爬行类、鸟类和哺乳类等系统发育和种群划分(Cecilia, 1994; Yu et al, 2000)。在本研究中,ND1和ND2基因中碱基A+T的平均含量分别为49.5%和49.8%,低于GC含量。单倍型多样型指数(Hd)和核苷酸多样性指数(Pi)作为衡量物种遗传多样性丰富度的重要指标,物种丰富度越高,在环境中的耐受性越强(陆键萍等, 2020)。本研究发现,

近年来,生物地理学成为鱼类分子群体遗传学的研究热点之一,在检测地理隔离对鱼类群体遗传结构的影响的研究中,线粒体DNA是比较理想的分子标记(祁得林等, 2008)。Iguchi等(2012)基于PCR-RFLP技术对日本4种








|
图 3 基于ND2基因构建的ML系统进化树(枝上数字为置信度值)
Fig.3 Maximum-likelihood Phylogenetic tree based on ND2 gene (The number on branch is confidence value)
SL:中国黄条 ;JSL:日本黄条 ;JSL:日本黄条 ;ASL:澳大利亚黄条 ;ASL:澳大利亚黄条 ;SQ:中国五条 ;SQ:中国五条 ;SD:中国高体 ;SD:中国高体 SL: Seriola lalandi from China; JSL: Seriola lalandi from Japan; ASL: Seriola lalandi from Australia; SQ: Seriola quinqueradiata from China; SD: Seriola dumerili from China
SL: Seriola lalandi from China; JSL: Seriola lalandi from Japan; ASL: Seriola lalandi from Australia; SQ: Seriola quinqueradiata from China; SD: Seriola dumerili from China
|
综上所述,本研究发现,ND1和ND2基因序列均可作为



BARRETT R D H, HEBERT P D N. Identifying spiders through DNA barcodes. Canadian Journal of Zoology, 2005, 83(3): 481-491 DOI:10.1139/z05-024 |
BI X X, GAO T X, XIAO Y S, et al. Sequence comparison of mtDNA 16S rRNA, CO Ⅰ and Cyt b gene fragments in four species of Gadidae fish. South China Fisheries Science, 2009, 5(3): 46-52 [毕潇潇, 高天翔, 肖永双, 等. 4种鳕鱼线粒体16S rRNA、CO Ⅰ和Cyt b基因片段序列的比较研究. 南方水产, 2009, 5(3): 46-52 DOI:10.3969/j.issn.1673-2227.2009.03.008] |
BOWEN B R, KREISER B R, MICKLE P F, et al. Phylogenetic relationships among North American Alosa species (Clupeidae). Journal of Fish Biology, 2008, 72(5): 1188-1201 DOI:10.1111/j.1095-8649.2007.01785.x |
CECILIA S. The evolution of mitochondrial DNA. Current Opinion in Genetics and Development, 1994, 4(6): 875-881 DOI:10.1016/0959-437X(94)90073-6 |
CHEN S H, OU Y J, LI J E. Mitochondrial DNA and its progresses in fish. Biotechnology Bulletin, 2011(3): 13-20 [陈四海, 区又君, 李加儿. 鱼类线粒体DNA及其研究进展. 生物技术通报, 2011(3): 13-20] |
CLAYTON D A. Transcription and replication of animal mitochondrial DNAs. International Review of Cytology, 1992, 141: 217-232 |
CUI A J. Study on population genetics of yellowtail kingfish. Master´ s Thesis of Shanghai Ocean University, 2020, 5-6 [崔爱君. 黄条  种群遗传特性研究. 上海海洋大学硕士研究生学位论文, 2020, 5-6] 种群遗传特性研究. 上海海洋大学硕士研究生学位论文, 2020, 5-6] |
GAO Z Y. Genetic diversity of wild populations of Channa argus in China. Master´ s Thesis of Jinan University, 2013, 20-36 [高志远. 中国野生乌鳢(Channa argus)遗传多样性分析. 暨南大学硕士研究生学位论文, 2013, 20-36] |
HEBERT P D N, STOECKLE M Y, ZEMLAK T S, et al. Identification of birds through DNA barcodes. PLoS Biology, 2004, 2(10): e312 DOI:10.1371/journal.pbio.0020312 |
IGUCHI J, TAKASHIMA Y, NAMIKOSHI A. Species identification method for marine products of Seriola and related specie. Fisheries Science, 2012, 78(1): 197-206 DOI:10.1007/s12562-011-0433-9 |
IRWIN D M, KOCHER T D, WILSON A C. Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution, 1991, 32(2): 128-144 DOI:10.1007/BF02515385 |
LIANG R S, LIANG Z G, FANG H H, et al. Molecular phylogenetic relationships of 16 sweetlips fishes based on the sequence of NADH dehydrogenase subunit 1. Haiyang Xuebao, 2014, 36(10): 25-32 [梁日深, 梁志刚, 方浩航, 等. 基于ND1基因序列的16种胡椒鲷鱼类分子系统进化关系研究. 海洋学报, 2014, 36(10): 25-32 DOI:10.3969/j.issn.0253-4193.2014.10.003] |
LIU X Z, XU Y J, LI R, et al. Analysis and evaluation of nutritional composition of the muscle of yellowtail kingfish (Seriola aureovittata). Progress in Fishery Sciences, 2017, 38(1): 128-135 [柳学周, 徐永江, 李荣, 等. 黄条  (Seriola aureovittata)肌肉营养组成分析与评价. 渔业科学进展, 2017, 38(1): 128-135] (Seriola aureovittata)肌肉营养组成分析与评价. 渔业科学进展, 2017, 38(1): 128-135] |
LU J P, YAO L, XIN H M, et al. Applicability analysis of mitochondrial CO Ⅰ, Cyt b and 16S rRNA genes in identification of six tuna species. Progress in Fishery Sciences, 2020, 41(5): 73-81 [陆键萍, 姚琳, 信红梅, 等. 线粒体CO Ⅰ、Cyt b和16S rRNA基因在6种金枪鱼鉴定中的适用性分析. 渔业科学进展, 2020, 41(5): 73-81] |
NWANI C D, BECKER S, BRAID H E, et al. DNA barcoding discriminates freshwater fishes from southeastern Nigeria and provides river system-level phylogeographic resolution within some species. Mitochondrial DNA: Journal of DNA Mapping, Sequencing and Analysis, 2011, 22(S1): 43-51 |
PREMACHANDRA H K A, TAKEUCHI Y, ADAM M, et al. Genomic DNA variation confirmed Seriola lalandi comprises three different populations in the Pacific, but with recent divergence. Scientific Reports, 2017, 7(1): 314-325 DOI:10.1038/s41598-017-00387-2 |
QI D L, CHAO Y, GUO S C, et al. Genetic structure of five Huanghe schizothoracin Schizopygopsis pylzovi populations based on mtDNA control region sequences. Journal of Zoology, 2008, 54(6): 972-980 [祁得林, 晁燕, 郭松长, 等. 黄河裸裂尻鱼五种群mtDNA控制区的遗传结构. 动物学报, 2008, 54(6): 972-980] |
QIAO H Y. Phylogeny of Schizothoracinae fishes and genetic structure of Pampus argenteus population based on mitochondrial DNA sequences. Master´ s Thesis of Shanghai Ocean University, 2014, 16-17 [乔慧莹. 基于线粒体基因序列分析裂腹鱼亚科的系统进化关系及中国沿海银鲳群体的遗传结构. 上海海洋大学硕士研究生学位论文, 2014, 16-17] |
SANGER O I, CARMONA J A, DOADRIO I. Evolutionary and biogeographical patterns within Iberian populations of the genus Squalius inferred from molecular data. Molecular Phylogenetics and Evolution, 2003, 29(1): 20-30 DOI:10.1016/S1055-7903(03)00088-5 |
SEPULVEDA F A, GONZALEZ M T. DNA barcoding evidence for the first recorded transmission of Neobenedenia sp.from wild fish species to Seriola lalandi cultured in an open recirculating system on the coast of Northern Chile. Aquaculture, 2019, 501: 239-246 |
SERB J M, BUHAY J E, LYDEARD C. Molecular systematics of the North American freshwater bivalve genus Quadrula (Unionidae: Ambleminae) based on mitochondrial ND1 sequences. Molecular Phylogenetics and Evolution, 2003, 28(1): 1-11 DOI:10.1016/S1055-7903(03)00026-5 |
TAKEYAMA H, CHOW S, TSUZUKI H, et al. Mitochondrial DNA sequence variation within and between tuna Thunnus species and its application to species identification. Journal of Fish Biology, 2001, 58(6): 1646-1657 DOI:10.1111/j.1095-8649.2001.tb02319.x |
WANG M, LIU H, HUANG H, et al. Identifying fish products in Shenzhen through DNA barcoding. Food Science, 2015, 36(20): 247-251 [王敏, 刘荭, 黄海, 等. DNA条形码技术在深圳鱼肉制品鉴定中的应用. 食品科学, 2015, 36(20): 247-251 DOI:10.7506/spkx1002-6630-201520048] |
WILSON A C, CANN R L, CARR S M, et al. Mitochondrial DNA and two perspectives on evolutionary genetics. Biology Journal of Society, 2010, 26(4): 375-400 |
YU N, ZHENG C L, ZHANG Y P, et al. Molecular systematics of pikas (genus Ochotona) inferred from mitochondrial DNA sequences. Molecular Phylogenetics and Evolution, 2000, 16(1): 85-95 |
YUAN J, ZHANG Q Z, LUO F. Mitochondrial DNA and its application to the molecular population genetics of fish. Ecological Science, 2008, 27(4): 272-276 [袁娟, 张其中, 罗芬. 鱼类线粒体DNA及其在分子群体遗传研究中的应用. 生态科学, 2008, 27(4): 272-276] |
ZHAO K. Mitochondrial DNA (mtDNA) of fish and application in molecular phylogenetic. Journal of Qinghai University (Natural Science), 2006, 24(2): 49-53 [赵凯. 鱼类线粒体DNA(mtDNA)及其在分子系统学中的应用. 青海大学学报(自然科学版), 2006, 24(2): 49-53] |
ZHENG D Y, GUO Y J, YANG T Y, et al. Genetic diversity analysis of Sillago japonica based on mitochondrial DNA ND2 gene. South China Fisheries Science, 2019, 15(5): 84-91 [郑德育, 郭易佳, 杨天燕, 等. 基于线粒体ND2基因序列的少鳞鱚遗传多样性研究. 南方水产科学, 2019, 15(5): 84-91] |
ZHENG W J, ZHU S H, ZHOU J X, et al. Molecular phylogenetic relationship of Carangidae based on partial sequence of mitochondrial 16S ribosomal RNA gene. Journal of Fisheries of China, 2008, 32(6): 847-854 [郑文娟, 朱世华, 邹记兴, 等. 基于16S rRNA部分序列探讨12种鲹科鱼类的分子系统进化关系. 水产学报, 2008, 32(6): 847-854] |