2. 福建省水产生物育种与健康养殖工程研究中心福建 厦门 361021;
3. 农业农村部东海海水健康与养殖重点实验室 福建 厦门 361021
2. Fujian Engineering Research Center of Aquatic Breeding and Healthy Aquaculture, Xiamen, Fujian 361021, China;
3. Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture and Rural Affairs, Xiamen, Fujian 361021, China
坛紫菜(Neoporphyra haitanensis)是我国沿海的一种大型经济海藻,年产量占全国紫菜总产量的75%以上,创造了可观的经济和社会效益(农业农村部渔业渔政管理局等, 2021),其适宜生长温度为16~24℃,最适生长温度约为21℃(曾呈奎等, 1985)。近年来,全球变暖导致福建、浙江等地的坛紫菜出现大面积烂苗现象,对沿海坛紫菜养殖业产生了巨大冲击,严重影响坛紫菜栽培产业的发展(宋武林, 2009)。因此,解析坛紫菜响应高温胁迫的分子机理,挖掘耐高温基因,指导坛紫菜耐高温品种选育,是从根本上解决当前坛紫菜高温烂苗的必经之路。
研究表明,植物在应答非生物胁迫时,可以通过转录因子控制下游靶基因以特定的强度在特定的时间和空间表达,进而调控植物自身的逆境胁迫应答过程。碱性亮氨酸拉链(basic region-leucine zipper, bZIP)家族转录因子作为植物中最庞大、最保守的转录因子家族之一,在植物响应高温、干旱、渗透等非生物胁迫中发挥着重要作用(Golldack et al, 2014)。bZIP转录因子家族含有2个结构域,即N末端中的碱性区域,可识别基因启动子上的保守序列与之结合;以及C末端中的亮氨酸拉链区域(Zulfiqar et al, 2016; E et al, 2014),用于行使激活和抑制功能(刘慧洁等, 2019)。bZIP已被证实可以通过结合功能基因或调节基因的启动子顺式元件来激活诱导下游基因的表达(Amorim et al, 2017; 孙耀国等, 2021)。例如,Li等(2020)研究发现,玉米(Zea mays) ZmbZIP60直接或间接激活诱导ZmHSF6B等抗逆基因的表达,从而调控玉米的耐热性;Zong等(2016)研究发现,水稻(Oryza sativa)的OsbZIP23基因在ABA介导的抗旱性中起中枢调控作用。迄今为止,已在拟南芥(Arabidopsis thaliana)、水稻、玉米中分别发现了127、89和216个bZIP转录因子(崔荣秀等, 2019)。但目前关于bZIP的研究主要集中于模式植物和部分大田作物,大型海藻中尚未有关于bZIP的报道。
本课题组前期研究发现,在高温胁迫下,坛紫菜的NhbZIP1基因显著上调表达,但其响应高温胁迫的分子机制尚不清楚。为此,本研究以坛紫菜全基因组和转录组数据为基础,对坛紫菜bZIP转录因子进行筛选和克隆,并分析其结构和表达模式。进一步将坛紫菜转录因子NhbZIP1基因转入莱茵衣藻中进行基因功能验证,为阐明bZIP调控坛紫菜抗逆的分子机制奠定理论基础。
1 材料与方法 1.1 实验材料实验材料选自集美大学坛紫菜种质改良及应用实验室种质资源库。培养条件:温度为(21.0±0.5)℃,光照强度为50~60 μmoL/(m2·s),光照周期为12 L︰12 D,每2 d更换一次新鲜Provasoli's enrichment solution (PES)叶状体培养液。PES母液配制:NaNO3 100 g、Na2HPO4·12H2O 20 g及微量元素,均定容至1000 mL。培养藻体长度至(15±2) cm,选取生长状态良好、无破损、无扭曲、平面光滑的藻体用于后续实验。
莱茵衣藻选自本实验室培养的细胞壁缺失株系“CC-400 cw15 mt+”。培养条件:温度为(25.0±0.5)℃,光照强度为50 μmoL/(m2·s),光照周期为14 L︰10 D,每7 d更换TAP培养基进行传代,培养至细胞密度为1~2×106个/mL用于后续实验(Yamano et al, 2013)。
1.2 总RNA分离纯化和cDNA合成采用E.Z.N.A植物RNA提取试剂盒(OMEGA,美国)提取坛紫菜和莱茵衣藻总RNA。通过Cary50紫外分光光度计(Varian, 美国)分别测定OD260 nm和OD280 nm值,根据计算结果初步判断RNA提取纯度及浓度,后续使用1%琼脂糖凝胶电泳检测RNA质量,判断RNA完整性和质量。
1.3 坛紫菜NhbZIP1的筛选及克隆根据课题组前期研究获得的坛紫菜基因组及转录组注释结果,在相关注释信息文件中以“bZIP”、“basic region-leucine zipper”为关键词进行搜索,将搜索获得的基因序列信息在NCBI中(https://www.ncbi.nlm.nih.gov)进行BLAST比对,最终筛选出1条注释结果为粳稻(Oryza sativa japonica Group) OsbZIP23基因的Gene000717。扩增体系总体积为25 μL:2×Mix (TaKaRa) 12.5 μL,dd H2O 9.5 μL,5′和3′引物各0.5 μL,cDNA模板2 μL。PCR扩增程序:预变性94℃ 30 s;变性98℃ 10 s,退火58.6℃ 30 s,延伸72℃ 2 min,共35个循环;最终延伸72℃ 10 min。将扩增产物回收纯化后进行测序验证,根据测序结果,获得NhbZIP1基因全长序列。
1.4 NhbZIP1生物信息学分析使用在线软件ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/)分析NhbZIP1基因核苷酸及氨基酸序列,预测分析开放阅读框位置,获得编码基因的起始密码子和终止密码子。使用在线软件SMART (http://smart.embl-heidelberg.de/)分析保守结构域。使用在线软件ExPASy (http://web.expasy.org/protparam/)分析NhbZIP1基因所编码蛋白质的一级结构。使用在线软件TMHMM (http://www.cbs.dtu.dk/services/TMHMM/)分析NhbZIP1基因所编码蛋白质是否存在跨膜结构域。使用磷酸化位点在线预测网站(http://www.cbs.dtu.dk/services/NetPhos/)和O-连接糖基化位点在线预测网站(http://www.cbs.dtu.dk/services/YinOYang/)分析NhbZIP1基因所编码蛋白质潜在的磷酸化和O-连接糖基化位点。使用在线软件WoLF PSORT (http://www.wolfpsort.org/)对NhbZIP1基因进行亚细胞定位预测。使用MEGA6软件的Neighbor-Joining法构建NhbZIP1系统进化树。
1.5 NhbZIP1莱茵衣藻表达载体构建及转化在目的基因起始密码子和终止密码子两端分别加上KpnⅠ和PstⅠ2个酶切位点碱基序列,使用KpnⅠ和PstⅠ内切酶对目的基因和莱茵衣藻表达载体pChlamy_3分别进行双酶切。双酶切体系:目的基因/ pChlamy_3载体2 μg,10×mol/L缓冲液2 μL,KpnⅠ内切酶1 μL,PstⅠ内切酶1 μL,用dd H2O补足体系。双酶切程序:37℃ 140 min,65℃ 10 min。双酶切后的目的基因和表达载体在16℃条件下过夜连接,将连接产物转化至感受态细胞E.coli DH5α中,挑选含有氨苄抗性的阳性单克隆菌落,进行PCR验证后扩繁培养,使用去内毒素质粒中量提取试剂盒(TIANGEN)提取质粒,获得含有目的基因的pChlamy_3莱茵衣藻表达载体。通过玻璃珠转化法(林键章等, 2021)将目的基因转化至莱茵衣藻中,筛选具有潮霉素抗性品系的单克隆衣藻进行扩繁培养,用于后续实验。
1.6 qRT-PCR分析NhbZIP1的表达水平本研究采用qRT-PCR技术检测NhbZIP1在32℃高温处理条件下不同时间点(0、5、10、15、30、60、180和360 min)的相对表达水平。根据克隆获得的NhbZIP1基因序列设计qRT-PCR引物(表 1),分别以NhUBC和β-tubulin基因作为坛紫菜和莱茵衣藻定量的内参基因。反应体系总体积为20 μL:2×SYBR green master mix (TaKaRa) 10 μL,dd H2O 6.8 μL,Rox Dye 0.4 μL,5′和3′引物各0.4 μL,cDNA模板2 μL。扩增程序:预变性95℃ 30 s;变性95℃ 5 s,退火及延伸58.6℃ 30 s,共40个循环。qRT-PCR扩增在Step One Plus型荧光定量PCR仪(ABI, 美国)上进行。
|
|
表 1 实验用到的引物序列 Tab.1 Primers used in this experiment |
野生型莱茵衣藻和转基因莱茵衣藻培养至对数生长期,通过Cary50紫外分光光度计(Varian, 美国)测定OD750 nm值约为0.3。将野生型和转基因莱茵衣藻同时置于32℃恒温培养箱中,控制其他相同条件不变。抽取样本通过外观表型观察及检测OD750 nm值来判断在不同时间处理下(0、24、48和72 h)其生物量变化差异。
1.8 转基因莱茵衣藻中抗逆相关基因的qRT-PCR分析采用qRT-PCR技术检测莱茵衣藻CAT、SOD抗氧化酶编码基因及HSP70A、HSP70B、HSP90A、HSP90C热激蛋白家族基因等相关抗逆基因在32℃高温处理条件下不同时间点(0、1、2、3 h)的相对表达水平变化。qRT-PCR扩增在Step One Plus型荧光定量PCR仪(ABI, 美国)上进行。
1.9 数据处理与分析所有实验处理均设置4个生物学重复。利用软件SPSS 23.0和Excel对实验数据结果进行统计分析,并采用单因素方差分析(one-way ANOVA)比较不同数据组间的差异,P < 0.05表示存在显著差异,P < 0.01表示差异极显著。用GpaphPad Prism 8.0软件作图呈现数据分析结果。
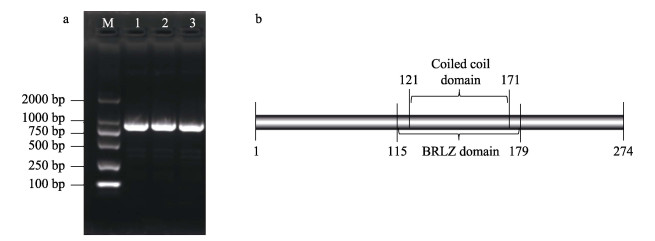
2 结果与分析 2.1 NhbZIP1的克隆及序列分析经PCR扩增获得一条长度约为1000 bp的产物(图 1a),经测序和BLAST分析比对,确定该基因为坛紫菜bZIP基因,命名为NhbZIP1。该基因开放阅读框起始密码子(ATG)到终止密码子(TAG)共825 bp,编码274个氨基酸,包含1个碱性区域亮氨酸拉链(basic region leucin zipper, BRLZ, 115~179 aa)结构域(图 1b)。该蛋白不存在信号肽及跨膜结构,定位于细胞核中。
|
图 1 NhbZIP1基因克隆及蛋白结构域分析 Fig.1 Cloning validation and protein domain analysis of NhbZIP1 gene a:NhbZIP1基因克隆产物电泳;b:NhbZIP1蛋白保守结构域 a: Agarose electrophoresis of PCR products of NhbZIP1 gene; b: Protein conservative domain of NhbZIP1 |
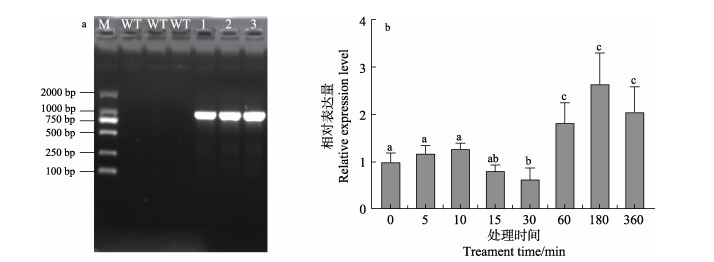
高温对坛紫菜NhbZIP1基因表达水平有显著影响,具体结果如下:在高温处理5 min时,NhbZIP1基因表达水平显著提高(P < 0.05),表达水平是对照组的1.6倍。在随后处理的60 min内,基因表达量逐渐降低,甚至降到初始水平以下,而在高温处理的180~360 min,基因表达量又显著提高,在高温处理360 min时表达水平约为初始水平的3.4倍(图 2)。

|
图 2 坛紫菜NhbZIP1基因在32℃高温胁迫下的表达模式 Fig.2 Expression pattern of NhbZIP1 gene in N. haitanensis under high temperature stress at 32℃ 不同上标字母表示数据间存在显著性差异(P < 0.05),下同。 Different superscript letters indicate significant differences between data (P < 0.05), the same as below. |
转基因实验结果显示,野生型莱茵衣藻未扩增出相应目的基因片段,而转基因莱茵衣藻均检测到阳性PCR产物(图 3a),表明已将坛紫菜NhbZIP1基因成功转化到莱茵衣藻中。选择第1个转基因品系(T1)用于后续实验分析。荧光定量PCR结果显示,高温处理30 min前,NhbZIP1基因表达量相对稳定,而在处理60 min时,基因表达水平显著提高(P < 0.05),表达水平是0 min时的1.8倍。在随后的高温胁迫下,基因表达量维持较高水平,在高温处理180 min时,表达水平约为初始水平的2.6倍(图 3b)。
|
图 3 转化藻株的筛选验证及NhbZIP1基因表达模式 Fig.3 Screening and validation of algal transformants and expression pattern of NhbZIP1 gene a:野生型莱茵衣藻和转基因莱茵衣藻目的基因PCR检测M为DL2000 Marker,WT:野生型莱茵衣藻,1~3:转基因莱茵衣藻,每个实验组各3个重复。b:高温32℃胁迫下NhbZIP1基因在莱茵衣藻中的相对表达水平 a: PCR detection of target gene of wild type C. reinhardtii and the algal transformants M: DNA Maker DL2000, WT: Wild type C. reinhardtii, 1~3: Genetically modified C. reinhardtii. There were three replicates in each group. b: Relative expression level of NhbZIP1 gene in C. reinhardtii under 32℃ high temperature stress |
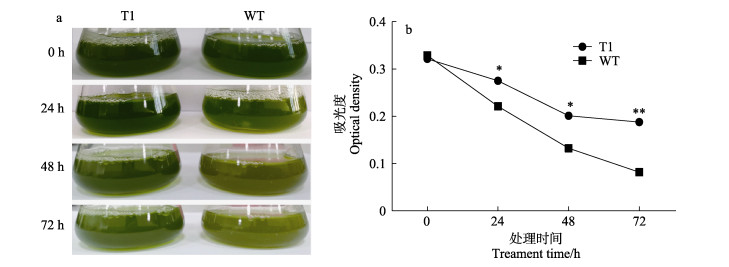
随着高温处理时间的增加,转基因莱茵衣藻和野生型莱茵衣藻的藻体都逐渐变黄(图 4a),表明在32℃高温胁迫下藻体死亡量逐步增加,但转基因莱茵衣藻存活率始终高于野生型(图 4b),且随着处理时间增加,差异越为显著。实验结果显示,转基因莱茵衣藻耐热性要高于野生型。
|
图 4 高温32℃胁迫下转基因(T1)和野生型(WT)莱茵衣藻的生物量变化 Fig.4 Biomass of transgenic C. reinhardtii overexpressing NhbZIP1 (T1) and wild type (WT) C.reinhardtii under 32℃ high temperature stress *表示差异显著,P < 0.05;**表示差异极显著,P < 0.01。下同。 * indicates significant difference at P < 0.05 level; ** indicates highly significant difference at P < 0.01 level. The same as below. |
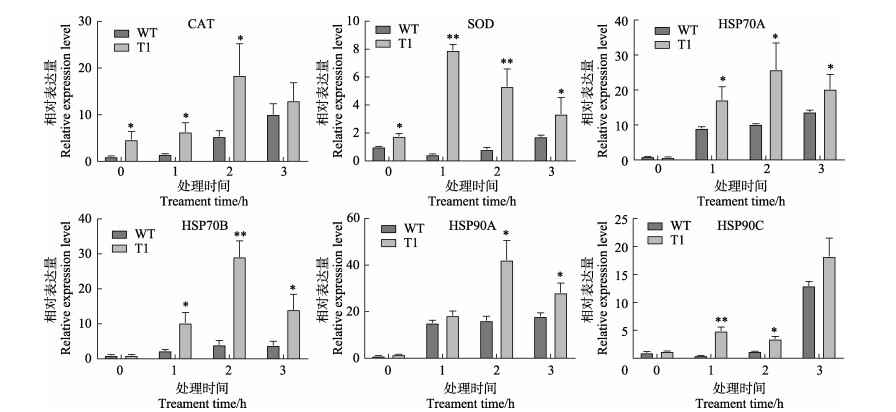
研究结果显示(图 5),抗氧化酶系统的CAT和SOD在高温处理的1~3 h中,转基因株系的转录水平均显著高于野生型。而热激蛋白家族基因也呈现类似现象,处理前期,基因表达量在转基因衣藻中呈现逐步上升趋势,HSP70A、HSP70B、HSP90A基因在转基因处理组中胁迫1~2 h表达量达到最高水平,转基因株系中基因表达量始终高于野生型。
|
图 5 32℃高温胁迫下转基因(T1)和野生型(WT)莱茵衣藻抗氧化酶和热激蛋白基因的相对转录水平 Fig.5 Relative transcript levels of genes encoding antioxidant enzymes and heat shock proteins in transgenic C. reinhardtii overexpressing NhbZIP1 (T1) and wild type (WT) C. reinhardtii under 32℃ high temperature stress |
本研究克隆获得了转录因子NhbZIP1基因,与前人研究对比发现,该基因也存在一个BRLZ结构,在121~171 aa之间是α卷曲螺旋结构;同时,该基因定位于细胞核中,符合转录因子的一般特征(邓冰等, 2021),表明NhbZIP1基因为bZIP家族转录因子。本研究发现,在高温处理下该基因显著上调表达,而在小麦(Geng et al, 2018)和水稻(Zong et al, 2020)的研究中也发现同样的现象,TabZIP60基因和OsbZIP23基因的表达量在高温和干旱胁迫前期也显著升高,推断NhbZIP1可以快速响应非生物胁迫以激活下游抗逆基因。这一结果也与课题组前期研究结果一致,Wang等(2018)研究发现,坛紫菜藻体在应答高温胁迫初期有一个应激反应,而后在转录水平进行重组,之后激活众多抗逆通路抵御长期高温胁迫。
3.2 NhbZIP1基因调控机制研究表明,bZIP激活HSP调控植物蛋白折叠稳态在其抵抗逆境胁迫中发挥着重要作用。玉米bZIP转录因子作为未折叠蛋白反应的诱导基因,可调控下游HSFTF13基因的表达来诱导HSPs合成,进一步通过激活连接未折叠蛋白反应和热激反应2种热胁迫响应系统(Li et al, 2020),促使玉米形成高温胁迫下的自我保护机制。同样,水稻中的bZIP转录因子可与OsHsfB2c基因启动子的反应元件相结合来调控HSF转录因子的表达,进而诱导体内HSPs的大量积累以抵御外界的非生物胁迫(李濯雪, 2015)。本课题组前期研究发现,HSPs参与维持胞内蛋白合成、折叠和清除,是坛紫菜适应逆境胁迫的关键基因(Wang et al, 2018、2019、2021; Chang et al, 2021)。进一步研究证实,bZIP可与下游含有AGCT核心基序的HSP相关基因结合,诱导其表达。而在坛紫菜HSPs基因启动子中同样含有ABRE等顺式元件结合基序(张宛晨等, 2021)。因此,坛紫菜NhbZIP1可能通过调节HSPs增加藻体的抗性。为了进一步验证这个推测,本研究将坛紫菜NhbZIP1基因转化至莱茵衣藻中进行功能验证。本研究发现,转基因莱茵衣藻中抗逆相关基因的转录水平显著高于野生型莱茵衣藻。同时,Wang等(2019)研究发现,坛紫菜PhHSP22基因可通过调控蛋白质动态平衡等途径增加转基因衣藻的耐高温能力。因此,推测NhbZIP1基因可能通过调控HSP的转录进而调控藻体应答高温胁迫过程。
除了维持蛋白质动态平衡,维持氧化还原动态平衡也是坛紫菜响应非生物胁迫的关键过程(张元等, 2011)。坛紫菜在进化和发育过程中形成了一套高效的活性氧清除机制,即抗氧化系统(张元等, 2011; 林颖辉等, 2018; 徐严等, 2018)。抗氧化酶在紫菜的抗氧化系统中起到不可替代的作用,例如超氧化物歧化酶(SOD)和过氧化氢酶(CAT)等(Wang et al, 2018)。Kranner等(2002)研究发现,过表达ThbZIP1基因可提高SOD等抗氧化酶的活性,进而加快活性氧的清除,以增强植株在逆境胁迫下的抗性。同样,拟南芥转CAbZIP基因植株也可通过调控CAT等抗氧化酶清除体内活性氧,进而提升转基因植株的抗旱性和抗盐性(曹红利等, 2012)。而本研究与上述研究结果相同,无论是正常条件下还是高温胁迫条件下,NhbZIP1转基因植株中CAT和SOD的转录水平均显著高于野生型。因此,坛紫菜NhbZIP1激活抗氧化酶编码基因也可能是机体耐高温的另一个重要原因。
4 结论本研究首次在坛紫菜中克隆获取了bZIP家族转录因子NhbZIP1基因,该基因可以快速响应高温胁迫。转NhbZIP1基因莱茵衣藻的耐热性显著高于野生型莱茵衣藻,在高温胁迫下,转基因株系的热激蛋白家族和抗氧化酶相关基因的表达水平均显著高于野生型。以上结果表明,NhbZIP1在坛紫菜应答高温胁迫中发挥着重要作用。
AMORIM LLB, DA FONSECA DOS SANTOS R, NETO JPB, et al. Transcription factors involved in plant resistance to pathogens. Current Protein and Peptide Science, 2017, 18(4): 335-351 DOI:10.2174/1389203717666160619185308 |
Bureau of Fisheries, Ministry of Agriculture and Rural Affairs, National Fisheries Technology Extension Center, China Society of Fisheries. China fishery statistical yearbook 2021. Beijing: China Agriculture Press, 2021 [农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会. 2021中国渔业统计年鉴. 北京: 中国农业出版社, 2021]
|
CAO H L, YUE C, WANG X C, et al. Advance in bZIP transcription factors related with plant stress resistance. Journal of Southern Agriculture, 2012, 43(8): 1094-1100 [曹红利, 岳川, 王新超, 等. bZIP转录因子与植物抗逆性研究进展. 南方农业学报, 2012, 43(8): 1094-1100] |
CHANG J, SHI J Z, LIN J Z, et al. Molecular mechanism underlying Pyropia haitanensis PhHsp22-mediated increase in the high-temperature tolerance of Chlamydomonas reinhardtii. Journal of Applied Phycology, 2021, 33(2): 1137-1148 DOI:10.1007/s10811-020-02351-6 |
CUI R X, ZHANG Y W, CHEN X Q, et al. The latest research progress on the stress responses of bZIP involved in plants. Biotechnology Bulletin, 2019, 35(2): 143-155 [崔荣秀, 张议文, 陈晓倩, 等. 植物bZIP参与胁迫应答调控的最新研究进展. 生物技术通报, 2019, 35(2): 143-155] |
DENG B, LIU Z Q, YUAN X W, et al. Identification of MYB transcription factor family members of Flammulina filiformis and analysis of their expression pattern during fruiting body development. Acta Edulis Fungi, 2021, 28(5): 1-11 [邓冰, 刘宗奇, 袁学文, 等. 金针菇MYB转录因子家族成员鉴定及其在子实体形成中的表达模式分析. 食用菌学报, 2021, 28(5): 1-11] |
E Z G, ZHANG Y P, ZHOU J H, et al. Roles of the bZIP gene family in rice. Genetics and Molecular Research, 2014, 13(2): 3025-3036 DOI:10.4238/2014.April.16.11 |
GENG X L, ZANG X S, LI H R, et al. Unconventional splicing of wheat TabZIP60 confers heat tolerance in transgenic Arabidopsis. Plant Science, 2018, 274: 252-260 DOI:10.1016/j.plantsci.2018.05.029 |
GOLLDACK D, LI C, MOHAN H, et al. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Frontiers in Plant Science, 2014, 5: 151 |
KRANNER I, BECKETT R P, WORNIK S, et al. Revival of a resurrection plant correlates with its antioxidant status. The Plant Journal, 2002, 31(1): 13-24 DOI:10.1046/j.1365-313X.2002.01329.x |
LI Z X, TANG J, SRIVASTAVA R, et al. The transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize. The Plant Cell, 2020, 32(11): 3559-3575 DOI:10.1105/tpc.20.00260 |
LI Z X. Screening of proteins binding to OsHsfB2c promoter by yeast one-hybrid system. Masterxs Thesis of Hunan Agricultural University, 2015 [李濯雪. 利用酵母单杂交方法对OsHsfB2c启动子结合蛋白的筛选. 湖南农业大学硕士研究生学位论文, 2015]
|
LIN J Z, WANG W L, XU Y, et al. The cDNA cloning and functional verification of the PhCUL1 gene from Pyropia haitanensis. Progress in Fishery Sciences, 2021, 42(1): 193-200 [林键章, 王文磊, 徐燕, 等. 坛紫菜泛素连接酶PhCUL1基因克隆与功能验证. 渔业科学进展, 2021, 42(1): 193-200] |
LIN Y H, WANG W L, XU Y, et al. Cloning and expression analysis of serine hydroxyl methyltransferase (SHMT) genes from Pyropia haitanensis. Progress in Fishery Sciences, 2018, 39(5): 122-129 [林颖辉, 王文磊, 徐燕, 等. 坛紫菜丝氨酸羟甲基转移酶基因的克隆及表达特征. 渔业科学进展, 2018, 39(5): 122-129] |
LIU H J, XU H, QIU W Y, et al. Roles of bZIP transcription factors in plant growth and development and abiotic stress response. Acta Agriculturae Zhejiangensis, 2019, 31(7): 1205-1214 [刘慧洁, 徐恒, 邱文怡, 等. bZIP转录因子在植物生长发育及非生物逆境响应的作用. 浙江农业学报, 2019, 31(7): 1205-1214] |
SONG W L. Analysis on rotten seedlings of Porphyra haitanensis and its prevention method. Journal of Fisheries Research, 2009(2): 72-75 [宋武林. 坛紫菜烂苗原因分析及预防对策. 福建水产, 2009(2): 72-75] |
SUN Y G, CAI T R, JI X Z, et al. Genome-wide bioinformatics analysis of bZIP gene family in Pyrus communis. Forestry and Ecological Sciences, 2021, 36(1): 24-34 [孙耀国, 蔡天润, 姬行舟, 等. 西洋梨全基因组bZIP基因家族生物信息学分析. 林业与生态科学, 2021, 36(1): 24-34] |
WANG W L, CHANG J, ZHENG H Y, et al. Full-length transcriptome sequences obtained by a combination of sequencing platforms applied to heat shock proteins and polyunsaturated fatty acids biosynthesis in Pyropia haitanensis. Journal of Applied Phycology, 2019, 31(2): 1483-1492 |
WANG W L, LIN J Z, CHANG J, et al. A RING type ubiquitin ligase PhCUL4 is involved in thermotolerance of Pyropia haitanensis. Algal Research, 2021, 59: 102448 |
WANG W L, TENG F, LIN Y H, et al. Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. PLoS One, 2018, 13(4): e0195842 |
XU Y, WANG W L, XU K, et al. Effect of different saline stress on physiological indexes of Pyropia haitanensis. Journal of Applied Oceanography, 2018, 37(3): 380-386 [徐严, 王文磊, 许凯, 等. 不同盐度胁迫对坛紫菜叶状体生理指标的影响. 应用海洋学学报, 2018, 37(3): 380-386] |
YAMANO T, IGUCHI H, FUKUZAWA H. Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. Journal of Bioscience and Bioengineering, 2013, 115(6): 691-694 |
ZENG C K, WANG S J, LIU S J. Marine algae cultivation. Shanghai: Shanghai Scientific and Technical Publishers, 1985 [曾呈奎, 王素娟, 刘思俭. 海藻栽培学. 上海: 上海科学技术出版社, 1985]
|
ZHANG W C, ZHU J, YI L F. Genome-wide identification and bioinformatics analysis of HSP20 gene family in Pyropia haitanensis. Journal of Southern Agriculture, 2021, 52(6): 1527-1535 [张宛晨, 朱静, 易乐飞. 坛紫菜HSP20基因家族成员的全基因组鉴定及生物信息学分析. 南方农业学报, 2021, 52(6): 1527-1535] |
ZHANG Y, XIE C T, CHEN C S, et al. Physiological responses of gametophytic blades of Porphyra haitanensis to rising temperature stresses. Journal of Fisheries of China, 2011, 35(3): 379-386 [张元, 谢潮添, 陈昌生, 等. 高温胁迫下坛紫菜叶状体的生理响应. 水产学报, 2011, 35(3): 379-386] |
ZONG W, TANG N, YANG J, et al. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiology, 2016, 171(4): 2810-2825 |
ZONG W, YANG J, FU J, et al. Synergistic regulation of drought-responsive genes by transcription factor OsbZIP23 and histone modification in rice. Journal of Integrative Plant Biology, 2020, 62(6): 723-729 |
ZULFIQAR A, SAMARA S S, IHSAN K, et al. Functions of plantxs bZIP transcription factors. Pakistan Journal of Agricultural Sciences, 2016, 53(2): 303-314 |



