2. 中国水产科学研究院黄海水产研究所青岛海洋科学与技术试点国家实验室海洋渔业科学与食物产出过程功能实验室 山东 青岛 266071
2. Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Laboratory for Marine Fisheries Science and Food Production Processes, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao 266071, China
急性肝胰腺坏死病(AHPND)最初被称为早期死亡综合征(EMS),是对虾养殖中最常见、最严重的疾病之一(Shen et al, 2021)。近年来,在中国、越南、马来西亚、菲律宾、泰国和墨西哥等国家都相继报道了AHPND的暴发,给全球虾类养殖业造成重大经济损失(贾丹等, 2018)。2014年以来,我国养殖对虾的AHPND发病率高达60%~80%,养殖产能降低、产量不稳定,使我国从对虾出口国变为进口国。据报道,可引起AHPND的主要致病原是副溶血弧菌(Vibrio parahaemolyticus) (Wang et al, 2015)。此外,哈维氏弧菌(Vibrio harveyi) (Kondo et al, 2015)、坎贝氏弧菌(Vibrio campbellii) (Ahn et al, 2017)、欧文氏弧菌(Vibrio owensii) (Liu et al, 2015)也可造成类似的病症,呈现明显的病原多样性。
副溶血弧菌是一种革兰氏阴性、嗜盐细菌,在世界各地的海洋和河口环境中广泛传播(Yang et al, 2019)。研究表明,不是所有的副溶血弧菌都能引起AHPND,经全基因测序及基因敲除发现,pirA和pirB是引起对虾AHPND的主要致病因子(Lee et al, 2015)。其中,pirB毒素主要决定该菌的致病力,pirA毒力相对较弱(Dangtip et al, 2015)。确切地说,AHPND的致病原应是染色体外毒力质粒pVA1中携带二元毒素pirAVp和pirBVp的副溶血弧菌的特定菌株(Powers et al, 2021)。其后,其他弧菌也被发现携带二元毒素基因pirAVp和pirBVp (Dong et al, 2017; Restrepo et al, 2018; Dhar et al, 2019)。进一步研究表明,毒力质粒pVA1还编码一个分离后杀伤系统以及一套接合转移和动员基因,可促进其在不同弧菌之间的传播(Yang et al, 2019)。因此,针对致病因子pirA和pirB设计特异性引物用于准确检测AHPND及精确防控疾病具有重要的科学价值和现实意义。
现阶段,AHPND病原菌分子检测的技术手段主要有PCR、定量PCR和环介导等温扩增技术(LAMP)等(Arunrut et al, 2016)。实时荧光定量PCR (quantitative real-time PCR, qPCR)与常规PCR相比,具有污染少、定量准确、实时监测、自动化程度高等优点,该技术已被应用于转基因检测(Michelini et al, 2008)、环境科学(Cummings et al, 2003)以及医学(Lee et al, 2004)等领域。然而,该技术具有耗时长、涉及多种仪器和试剂、对人员的专业技能和经验要求高等特点。Kongrueng等(2015)设计的2组引物(LAMP-a2和LAMP-a3)在LAMP检测中用于特异识别引起AHPND的副溶血弧菌。上述方法虽然灵敏度高,但假阳性问题比较严重。微流控芯片检测技术允许多个单元的灵活组合并具有整体可控性,因此,包括样品制备和检测在内的一些步骤可以集成到单个芯片中。由于芯片是微米级甚至纳米级的,它具有高比表面积、高扩散系数、快速传热等特点(Zhao et al, 2019)。值得注意的是,将qPCR技术与微流控芯片技术相结合,与普通PCR相比,微流控荧光定量PCR技术具有实现高灵敏、多通道、自动化和现场快速检测的潜力。
在水产领域,微流控检测技术研究相对较少,本实验室在前期研究中,已经针对鱼类美人鱼发光杆菌美人鱼亚种(Photobacterium damselae subsp. Damselae)开展了微流控快速检测技术的研究(Zhang et al, 2021)。Sirikharin等(2015)研究表明,尽管毒力质粒编码的二元基因pirAVp和pirBVp是AHPND的主要致病因素,但毒力基因toxR和toxA在副溶血弧菌中普遍存在,并在对虾感染过程中通过16S rRNA基因的遗传多样性发挥重要作用。基于此,针对pirA和pirB基因设计特异性引物,通过优化反应体系和反应条件,本研究建立可用于现场检测的双重微流控荧光定量快速检测方法,为早期识别疾病风险和快速治疗提供技术保障。
1 材料与方法 1.1 实验菌株实验中使用的病原菌包括含有pirA和pirB的副溶血弧菌VpAHPND (标记为Vp1001),不含有pirA和pirB的副溶血弧菌(标记为F3-7)。此外,还包括实验室保藏的其他7种病原菌(表 1),用于特异性检验分析。
|
|
表 1 实验所用菌株 Tab.1 Strains used in the experiment |
本研究根据GenBank中登录的pirA和pirB基因的保守序列设计引物,再通过NCBI中Blast验证引物的特异性。设计完成的引物序列交由生工生物工程(上海)股份有限公司合成,引物序列见表 2。以VpAHPND作为检测目标菌,非致AHPND的副溶血弧菌(非VpAHPND)、哈维氏弧菌和大菱鲆弧菌等其他8株细菌作为对照菌,无菌水作为空白对照,使用实验室实时荧光定量PCR仪进行扩增,验证各引物的特异性。反应体系:2×Taq Pro Universal SYBR qPCR Master Mix 10 μL,正/反引物各0.4 μL,DNA模板2 μL,去离子水7.2 μL。反应条件:95 ℃ 30 s,95 ℃ 5 s,退火温度(58~63 ℃) 30 s,95 ℃ 15 s,进行40次循环,并依据扩增效果最终确定最佳退火温度及反应条件。
|
|
表 2 实验所用引物 Tab.2 Primers used in the experiment |
以VpAHPND的DNA为模板,用设计的pirA和pirB的2对引物分别进行常规PCR扩增,扩增产物切胶回收后连接至pMD-19T载体,转化至大肠杆菌(Escherichia coli) DH-5α感受态细胞,增菌培养,进行菌液PCR扩增后测序筛选出阳性菌作为标准品。将VpAHPND已构建的标准品增菌培养后提取质粒,按10倍梯度稀释6个梯度分别进行qPCR扩增并绘制标准曲线。
1.4 双重微流控荧光定量PCR灵敏度实验将构建的pirA和pirB质粒标准品分别按10倍梯度稀释为109~100 copies/μL。将稀释后的标准品分别作为模板进行qPCR扩增,每个梯度设3个平行,验证其灵敏度。
1.5 双重微流控荧光定量PCR反应条件的优化用含有pirA和pirB菌株的DNA作为模板,比较单重微流控荧光定量PCR的退火温度,分别在反应温度为58~63 ℃下进行扩增,最终筛选出2对引物最佳的共同退火温度。同时,分别设反应循环数为25、30、35和40,验证最佳反应循环数。
1.6 双重微流控荧光定量PCR芯片集成以VpAHPND和非VpAHPND的DNA作为模板,用实验室设计的pirA和pirB引物、toxR(徐媛媛等, 2022)和pirA (Han et al, 2015)于微流控荧光定量PCR上集成,验证本研究设计的2对引物在该仪器上应用的可行性以及准确性。
1.7 人工感染人工感染实验所用凡纳滨对虾(Litopenaeus vannamei)(体长为5~8 cm,平均体重为3.3 g)均购自山东省青岛市某养殖场。将健康对虾在水族实验室水桶中暂养1周,养殖用水经消毒、过滤、沉淀和曝气,实验期间连续充气,并保持水温为(27±2) ℃,盐度为20。攻毒实验采取浸浴和注射2种方式进行,实验分为7组,即VpAHPND浸浴和注射2组,非VpAHPND浸浴和注射2组,DH5α浸浴和注射2组,空白对照1组。挑选个体大小均匀、健康的对虾,每桶10尾。
F3-7和Vp1001两株实验菌分别在TSB培养基上进行培养,28 ℃恒温培养18~24 h,挑取单菌落培养于含有1.5% NaCl的TSB液体培养基中,28 ℃下恒温培养过夜,离心重悬后,在浸浴组养殖水体中加入预先制备好的菌液,使菌含量的终浓度为106 CFU/mL。在注射组中,注射30 μL菌液(浓度为106 CFU/mL)到对虾肌肉中。空白对照组只用养殖水饲养,实验期间不投喂。
1.8 不同对虾组织pirAB定量检测分别于攻毒后第2、6和12小时采集对照组与实验组的水样,并随机捞取2尾凡纳滨对虾,对肌肉、肝胰腺、鳃丝和肠道4个组织进行取样处理。将组织用剪刀剪碎并充分混匀,按照DNA提取试剂盒(TIANGEN)说明书步骤提取组织DNA。用提取好的DNA,在已经优化好的反应条件下进行多重微流控荧光定量PCR扩增,验证检测效果。
2 结果与分析 2.1 致病因子pirA和pirB引物特异性验证用预实验最终确定的退火温度(60 ℃),用本研究设计的pirA和pirB两对引物分别对VpAHPND菌株、非VpAHPND菌株、哈维氏弧菌、美人鱼发光杆菌美人鱼亚种、大菱鲆弧菌、鳗弧菌、灿烂弧菌、嗜环弧菌和轮虫弧菌同时进行qPCR检测。实验结果显示,只有VpAHPND菌株(菌株Vp1001)成功扩增获得pirA和pirB两个基因(图 1),而其他的病原菌检测结果均为阴性。结果表明,本研究所设计的pirA和pirB两对引物可以特异性检测出含有pirA和pirB的病原菌,而对其他不含有pirA和pirB的病原菌无扩增,满足特异性检测的需要。

|
图 1 pirA和pirB引物特异性验证 Fig.1 Specificity validation of pirA and pirB primer a: pirA; b: pirB |
将含有致病因子pirA和pirB的2个质粒标准品分别按10倍梯度稀释成6个梯度,进行qPCR扩增,并绘制标准曲线。结果如图 2a所示,pirA的标准曲线在5.43×109~5.43×104 copies/μL浓度范围内有良好的线性相关性(y= –3.145x+6.63,R2=0.999)。如图 2b所示,pirB的标准曲线在4.31×109~4.31×104 copies/μL浓度范围内有良好的线性相关性(y= –3.015x+5.45,R2=0.999)。

|
图 2 构建成功pirA和pirB的质粒标准品扩增曲线和标准曲线 Fig.2 Amplification curve and standard curve of the plasmid standard for the successful construction of pirA and pirB a: pirA; b: pirB; 1: 109 copies/μL; 2: 108 copies/μL; 3: 107 copies/μL; 4: 106 copies/μL; 5: 105 copies/μL; 6: 104 copies/μL |
将含有目的基因靶标的标准品连续稀释10倍作为模板,进行3个重复的荧光定量PCR,以去离子水进行阴性对照。pirA标准品DNA模板在101和100 copies/反应时,3个平行实验的Ct值分别为31.11、31.12、31.24和32.25、32.38、32.29;pirB标准品DNA模板在101 copies/反应时,3个平行实验的Ct值分别为31.96、31.99、32.15,且都有明显的扩增曲线。在实际应用中,为保证检测结果的可靠性和准确性,常以Ct值35.00为界限,当Ct值≤35.00且有扩增曲线,判定为阳性。因此,pirA和pirB的最低检测限分别为5.43×100和4.31×101 copies/μL (图 3)。

|
图 3 pirA和pirB荧光定量PCR灵敏度实验 Fig.3 Sensitivity test of pirA and pirB fluorescence quantitative PCR a: pirA; b: pirB; 1~10: 109, 108, 107, 106, 105, 104, 103, 102, 101 and 100 copies/μL |
优化致病因子pirA和pirB双重微流控荧光定量PCR扩增条件:以制备好的pirA和pirB质粒标准品分别作为模板,将荧光定量PCR体系分别在58~63 ℃下进行扩增,对扩增曲线进行分析。最终确定引物浓度为1 μmol/L,最佳反应温度为60 ℃,反应循环数为30,检测时间为26 min左右。
以VpAHPND和非VpAHPND的DNA作为模板,用本文设计的2对引物(pirA和pirB)和引物toxR于微流控荧光定量PCR上集成,验证3对引物在该仪器上应用的可行性;同时引用Han等(2005)的引物验证本研究设计引物的准确性。结果显示,共有5条扩增曲线,分别是非VpAHPND组的toxR 1条扩增曲线,VpAHPND组的pirA、pirB和toxR 3条扩增曲线,pirA (Han et al, 2005) 1条扩增曲线,非VpAHPND组pirA、pirB和空白对照组均未产生扩增曲线(图 4)。
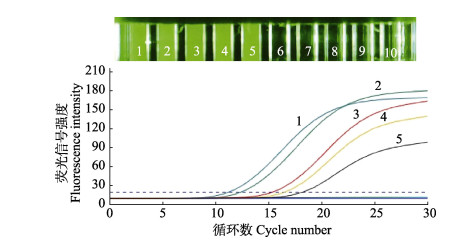
|
图 4 微流控集成图 Fig.4 Microfluidic integration diagram 1:VpAHPND pirA;2: VpAHPND pirB;3:非VpAHPND toxR (Non-VpAHPND toxR);4:VpAHPND toxR;5:pirA (Han et al, 2005);6:非VpAHPND pirA (Non-VpAHPND pirA);7:非VpAHPND pirB (Non-VpAHPND pirB);8:ddH2O |
选取凡纳滨对虾为实验对象,应用已建立的方法对7个实验组的水样和4个组织部位(鳃丝、肝胰腺、肌肉和肠道)进行样品检测,与实验室常规实时荧光定量PCR进行同步检测作对比。实验中,非VpAHPND组、大肠杆菌阳性对照组和空白组的样本检测结果均为阴性,VpAHPND组在对虾感染AHPND过程中不同感染方式的检测结果不同,在不同时间段各个组织先后感染程度也不相同(表 3、表 4)。
|
|
表 3 基于pirA和pirB微流控荧光定量PCR的应用效果对比 Tab.3 Comparison of application effects of microfluidic fluorescence quantitative PCR based on pirA and pirB |
|
|
表 4 基于toxR微流控荧光定量PCR的应用效果对比 Tab.4 Comparison of application effects of microfluidic fluorescence quantitative PCR based on toxR |
pirA基因在注射的实验组中,水样检测结果均为阴性,浸浴实验组水样均为阳性。且在注射实验组中,2~12 h时所取对虾的4个组织样品的检测结果均为阳性。在浸浴实验组中有着不同实验结果,2 h时,肝胰腺和肠道检测为阴性,鳃和肌肉为阳性;6 h时,只有肝胰腺为阴性,其他样品均为阳性;12 h时,全部组织均为阳性。
pirB基因的检测结果与注射感染pirA时的检测结果相同,与浸浴感染pirA时有所不同。在浸浴2 h时,只有肠道为阴性,其他样品均为阳性;6 h和12 h的全部样品均为阳性。
VpAHPND和非VpAHPND组的toxR基因的检测结果相同。浸浴组中,2 h时水样检测为阳性,4个组织检测结果均为阴性;6 h和12 h的所有样品均为阳性。注射组中,除了水样检测为阴性外,所有时间段的其他样品均为阳性。
在菌株检测水平,pirA的灵敏度优于pirB,但在人工感染实践中pirB比pirA有更高的灵敏度,因此,实验中的结果应用于生产实践时需结合宿主做进一步验证。同时,以上检测结果表明,致病因子pirA和pirB均具有较高的检出率与检测灵敏度,适用于AHPND致病原的检测。
3 讨论近年来,由弧菌引起的AHPND感染对虾比较频繁,该病可使对虾在感染后较短时间内大规模死亡,自2009年以来已给全球对虾养殖业造成430亿美元的损失(Kumar et al, 2021)。在我国,流行病学研究发现,该疾病致死率高、传播范围广,患病对虾通常表现为肝胰腺肿大,后期逐渐萎缩,其颜色从黄褐色、红褐色到浅褐色甚至白色,呈现厌食、空肠、空胃状态,甲壳变软、不易褪壳,池水常常漂浮白便。一般5~7 d内出现大批死亡,其死亡率高达80%以上(Chen et al, 2022)。在养殖实践中,养殖者往往通过肉眼观察发病后期所体现出的症状来判断疾病的发生,延误了治疗的最佳时期,最终导致对虾大批死亡。因此,以精准、灵敏、快速的现场检测手段,建立一种能够诊断AHPND的早期感染态,是及早认知疾病发生风险和快速进行治疗的重要措施。
从诊断技术发展进程来看,临床体征和组织病理学是早期检测AHPND的方法(Mai et al, 2021)。自2014年起,有学者建立了用于检测AHPND急性或终末期感染的PCR和反转录PCR (RT-PCR)方法(Dangtip et al, 2015)。尽管PCR的方法具有高灵敏性和精确性,但花费时间长,操作精细,需要专业人员完成检测。随着pirA和pirB二元毒素基因的发现,Cruz-Flores等(2019)设计了双重TaqMan实时荧光定量PCR检测方法,可同时检测pirA和pirB,检测时间约为27 min,但该方法对操作人员要求较高,也不能进行现场检测。Arunrut等(2016)基于LAMP,结合使用DNA功能化、ssDNA标记的纳米金探针(AuNP),实现对AHPND阳性扩增产物的可视化,总检测时间为50 min,虽然能实现现场检测,但检测时间较长。Mai等(2021)针对pirA和pirB致病基因建立了等温重组酶聚合酶扩增(RPA)检测方法,完成检测AHPND的时间为70 min,检测时间较长。陈蒙蒙等(2018)采用qPCR的方法检测AHPND,灵敏度为101 copies/µL,而本研究中基于pirA的微流控荧光定量PCR灵敏度更高(5.43×100 copies/µL)。微流控芯片检测技术具有体积小、样品和试剂消耗量少、检测速度快、兼具微型化和可集成化等优点,已成为检测技术关注的新热点。本研究针对AHPND建立了二元毒素编码基因pirA和pirB的微流控荧光定量PCR的检测方法,结果显示,微流控荧光定量PCR检测方法对致病因子pirA和pirB具有良好的特异性,能准确检测出VpAHPND,非VpAHPND以及对照组均未出现荧光信号。对pirA和pirB目的片段构建标准曲线发现,pirA的标准曲线在5.43×109~5.43×104 copies/μL浓度范围内有良好的线性相关性(y= –3.145x+6.63, R2=0.999),pirB的标准曲线在4.31×109~4.31×104 copies/μL浓度范围内有良好的线性相关性(y= –3.015x+5.45, R2=0.999)。2个基因在相同体系和条件下在同一芯片进行集成化,反应时间为26 min左右,满足了快速、灵敏、高通量、一体化集成的现场检测的实际需求。
Kumar等(2021)研究指出,除了pirABVP毒素,AHPND相关菌株还有其他特定的毒力因子,可能与AHPND引起的细菌的毒力和疾病有关。本研究中的人工感染实验采用注射和浸浴2种方式进行,在不同时间段取样,对所建立的方法进行临床验证并且比较其在对虾不同组织中检测的效果,可以更好地分析病原菌的侵染途径。结果发现,以注射为感染方式的实验组,除水样检测为阴性外,各个时间段的组织均为阳性。以浸浴为感染方式的实验组,不同时间段和不同基因检测的结果不同。从感染方式来看,采用注射方法,细菌2 h就可以到达各组织;而采用浸浴感染方法,浸浴6 h肝胰腺还未能检测到靶标基因,说明注射侵染组织速度快于浸浴感染方式。从浸浴感染的3个基因比较结果来看,pirB只有在2 h肠道检测为阴性,其他时间的组织都为阳性;pirA在2 h的肝胰腺和肠道检测为阴性,6 h只有肠道为阴性,12 h均为阳性;toxR在2 h各组织检测都为阴性,说明pirB基因侵染速度最快,pirA基因次之,toxR基因最慢。从组织被侵染速度来看,在2 h时,鳃丝和肌肉中均检测到pirA和pirB,被侵染速度最快。陈蒙蒙等(2018)研究表明,对虾的鳃丝比表面积大,是病原菌侵入的通道,并直通虾胃及肝胰腺,更可能成为病原菌直接入侵的通道。与本研究中pirA在浸浴2 h时肝胰腺和肠道的检测结果为阴性、鳃丝为阳性的结果相似。Lai等(2015)对人工感染对虾进行取样,6 h内对虾肝胰脏组织明显脱落,并检测出高浓度的pirB,感染18 h后才检出pirA,表明pirB的侵染速度快于pirA。这与本研究pirB比pirA基因检出率高的结果是一致的。综上,由于pirB检出率最高,鳃丝和肌肉被侵染速度最快,因此,以pirB为引物、以鳃丝或肌肉为靶组织的检测策略更适用于AHPND的现场快速检测。
综上所述,本研究建立了一种具有高灵敏度和特异性强的AHPND微流控荧光定量PCR检测方法,该方法对pirA的最低检测限可达到5.43×100 copies/μL,对pirB的最低检测限可达到4.31×101 copies/μL,具有较高灵敏度,检测时间为26 min左右,可以快速、准确地确认携带pirA和pirB的致病弧菌,为及早认知疾病发生风险和快速进行治疗提供技术保障。此外,本研究使用的仪器具有良好的便携性便于在养殖现场操作,并且检测反应时间短,对于AHPND的现场快速诊断具有良好的开发应用前景。
AHN Y S, PIAMSOMBOON P, TANG K F J, et al. Complete genome sequence of acute hepatopancreatic necrosis disease-causing Vibrio campbellii LA16-V1, isolated from Penaeus vannamei cultured in a Latin American country. Genome Announcements, 2017, 5(37): e01011-17 |
ARUNRUT N, KAMPEERA J, SIRITHAMMAJAK S, et al. Sensitive visual detection of AHPND bacteria using loop-mediated isothermal amplification combined with dna-functionalized gold nanoparticles as probes. PLoS One, 2016, 11(3): e0151769 DOI:10.1371/journal.pone.0151769 |
CHEN M M, DONG X, QIU L, et al. Quantitative analysis of acute hepatopancreatic necrosis disease causing Vibrio parahaemolyticus (VpAHPND) in infected Litopenaeus vannamei. Progress in Fishery Sciences, 2018, 39(4): 93-100 [陈蒙蒙, 董宣, 邱亮, 等. 凡纳滨对虾感染致急性肝胰腺坏死病副溶血弧菌(VpAHPND)的定量分析. 渔业科学进展, 2018, 39(4): 93-100] |
CHEN Y L, KUMAR R, LIU C H, et al. Litopenaeus vannamei peritrophin interacts with WSSV and AHPND-causing V. parahaemolyticus to regulate disease pathogenesis. Fish and Shellfish Immunology, 2022, 126: 271-282 DOI:10.1016/j.fsi.2022.05.035 |
CRUZ-FLORES R, MAI H N, DHAR A K. Multiplex SYBR Green and duplex TaqMan real-time PCR assays for the detection of photorhabdus insect-related (Pir) toxin genes pirA and pirB. Molecular and Cellular Probes, 2019, 43: 20-28 DOI:10.1016/j.mcp.2018.12.004 |
CUMMINGS D E, SNOEYENBOS-WEST O L, NEWBY D T, et al. Diversity of Geobacteraceae species inhabiting metal-polluted freshwater lake sediments ascertained by 16S rDNA analyses. Microbial Ecology, 2003, 46(2): 257-269 DOI:10.1007/s00248-005-8002-3 |
DANGTIP S, SIRIKHARIN R, SANGUANRUT P, et al. AP4 method for two-tube nested PCR detection of AHPND isolates of Vibrio parahaemolyticus. Aquaculture Reports, 2015, 2: 158-162 DOI:10.1016/j.aqrep.2015.10.002 |
DHAR A K, PIAMSOMBOON P, ARANGUREN CARO L F, et al. First report of acute hepatopancreatic necrosis disease (AHPND) occurring in the USA. Diseases of Aquatic Organisms, 2019, 132(3): 241-247 DOI:10.3354/dao03330 |
DONG X, WANG H, XIE G, et al. An isolate of Vibrio campbellii carrying the pirVP gene causes acute hepatopancreatic necrosis disease. Emerging Microbes and Infections, 2017, 6(1): e2 |
HAN J E, TANG K F J, PANTOJA C R, et al. qPCR assay for detecting and quantifying a virulence plasmid in acute hepatopancreatic necrosis disease (AHPND) due to pathogenic Vibrio parahaemolyticus. Aquaculture, 2015, 442: 12-15 DOI:10.1016/j.aquaculture.2015.02.024 |
JIA D, SHI C Y, HUANG J, et al. Identification and pathogenicity analysis of bacterial pathogen associated with acute hepatopancreatic necrosis disease (AHPND) in the Pacific shrimp Litopenaeus vannamei. Progress in Fishery Sciences, 2018, 39(3): 103-111 [贾丹, 史成银, 黄倢, 等. 凡纳滨对虾急性肝胰腺坏死病(AHPND)病原分离鉴定及其致病性分析. 渔业科学进展, 2018, 39(3): 103-111] |
KONDO H, VAN P T, DANG L T, et al. Draft genome sequence of non-Vibrio parahaemolyticus acute hepatopancreatic necrosis disease strain KC13.17.5, isolated from diseased shrimp in Vietnam. Genome Announcements, 2015, 3(5): e00978-15 |
KONGRUENG J, TANSILA N, MITRAPARP-ARTHORN P, et al. LAMP assay to detect Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease in shrimp. Aquaculture International, 2015, 23(5): 1179-1188 DOI:10.1007/s10499-014-9874-3 |
KUMAR V, ROY S, BEHERA B K, et al. Acute hepatopancreatic necrosis disease (AHPND): Virulence, pathogenesis and mitigation strategies in shrimp aquaculture. Toxins, 2021, 13(8): 524 DOI:10.3390/toxins13080524 |
LAI H C, NG T H, ANDO M, et al. Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish and Shellfish Immunology, 2015, 47(2): 1006-1014 DOI:10.1016/j.fsi.2015.11.008 |
LEE C T, CHEN I T, YANG Y T, et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(34): 10798-10803 DOI:10.1073/pnas.1503129112 |
LEE C W, SUAREZ D L. Application of real-time RT-PCR for the quantitation and competitive replication study of H5 and H7 subtype avian influenza virus. Journal of Virological Methods, 2004, 119(2): 151-158 DOI:10.1016/j.jviromet.2004.03.014 |
LIU L, XIAO J, XIA X, et al. Draft genome sequence of Vibrio owensii strain SH-14, which causes shrimp acute hepatopancreatic necrosis disease. Genome Announcements, 2015, 3(6): e01395-15 |
MAI H N, ARANGUREN CARO L F, CRUZ-FLORES R, et al. Development of a recombinase polymerase amplification (RPA) assay for acute hepatopancreatic necrosis disease (AHPND) detection in Pacific white shrimp (Penaeus vannamei). Molecular and Cellular Probes, 2021, 57: 101710 DOI:10.1016/j.mcp.2021.101710 |
MICHELINI E, SIMONI P, CEVENINI L, et al. New trends in bioanalytical tools for the detection of genetically modified organisms: An update. Analytical and Bioanalytical Chemistry, 2008, 392(3): 355-367 DOI:10.1007/s00216-008-2193-7 |
POWERS Q M, ARANGUREN F L, FITZSIMMONS K M, et al. Crayfish (Cherax quadricarinatus) susceptibility to acute hepatopancreatic necrosis disease (AHPND). Journal of Invertebrate Pathology, 2021, 186: 107554 DOI:10.1016/j.jip.2021.107554 |
RESTREPO L, BAYOT B, ARCINIEGAS S, et al. PirVP genes causing AHPND identified in a new Vibrio species (Vibrio punensis) within the commensal Orientalis clade. Scientific Reports, 2018, 8(1): 13080 DOI:10.1038/s41598-018-30903-x |
SHEN H, SONG T, LU J, et al. Shrimp AHPND causing Vibrio anguillarum infection: Quantitative diagnosis and identifying antagonistic bacteria. Marine Biotechnology, 2021, 23(6): 964-975 DOI:10.1007/s10126-021-10079-8 |
SIRIKHARIN R, TAENGCHAIYAPHUM S, SANGUANRUT P, et al. Characterization and PCR detection of binary, pir-like toxins from Vibrio parahaemolyticus isolates that cause acute hepatopancreatic necrosis disease (AHPND) in shrimp. PLoS One, 2015, 10(5): e0126987 DOI:10.1371/journal.pone.0126987 |
WANG R, ZHONG Y, GU X, et al. The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Frontiers in Microbiology, 2015, 6: 144 |
XU YY, YU YX, WANG YG, et al. Establishment of multiple microfluidic fluorescence quantitative PCR detection technology for five main mariculture bacterial pathogens. Progress in Fishery Sciences, 2023, 44(3): 222-234 [徐媛媛, 于永翔, 王印庚, 等. 5种主要海水养殖病原菌多重微流控荧光定量PCR快速检测技术的建立. 渔业科学进展, 2023, 44(3): 222-234] |
YANG Q, DONG X, XIE G, et al. Comparative genomic analysis unravels the transmission pattern and intra-species divergence of acute hepatopancreatic necrosis disease (AHPND)-causing Vibrio parahaemolyticus strains. Molecular Genetics and Genomics, 2019, 294(4): 1007-1022 DOI:10.1007/s00438-019-01559-7 |
ZHANG Z, YU Y, CHEN J, et al. Development of a microfluidics-based quantitative real-time PCR to rapidly identify Photobacterium damselae subsp. damselae with different pathogenicity by detecting the presence of mcp or dly gene. Journal of Ocean University of China, 2021, 20(2): 445-453 |
ZHAO X, LI M, LIU Y. Microfluidic-based approaches for foodborne pathogen detection. Microorganisms, 2019, 7(10): 381 DOI:10.3390/microorganisms7100381 |



