2. 南方海洋科学与工程广东省实验室(湛江) 广东 湛江 524025
2. Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), Zhanjiang 524025, China
军曹鱼(Rachycentron canadum)俗称海鲡、海龙,是大型肉食性暖水鱼类,主要分布于印度洋、西太平洋和大西洋沿岸,在我国的南海、东海及黄海均有分布(勾效伟等, 2007)。军曹鱼因其肉质细嫩、味道鲜美、营养价值高而深受消费者喜爱;其在营养成分上富含多种不饱和脂肪酸及微量元素,具有一定的药用价值;加之军曹鱼具有生长速度快、抗病力强等优点,因而成为我国最具养殖前景的海水鱼类之一(李刘冬等, 2002)。近年来,国内外学者针对军曹鱼的生长发育特征(邝杰华等, 2021; 毛非凡等, 2022)、繁殖生物学特征(邝杰华等, 2021)、营养需求(麻永财等, 2012)、人工繁育及苗种培育(陈刚等, 2004; 刘海娟等, 2012)等开展了大量研究。目前,关于军曹鱼遗传多样性水平的研究报道较少。王中铎等(2010、2011)利用简单重复序列(simple sequence repeat, SSR)分子标记和扩增片段长度多态分析技术(AFLP)对军曹鱼养殖群体的遗传多样性分析显示,选取的各养殖群体遗传多样性水平较高,其群体遗传分化水平与野生群体差异不大。李伟强等(2020)利用12个SSR标记对广东、海南等5个军曹鱼养殖群体的遗传多样性分析结果显示,各群体的遗传多样性水平较10年前均有所下降。由此推测,长期人工繁育可能会导致军曹鱼不同养殖群体间基因交流减少,群体遗传多样性水平降低。因此,加强军曹鱼种质资源保护与利用,对于其增养殖业的健康发展具有重要意义。
SSR分子标记也称为微卫星DNA序列标记,其最常见的重复单元类型为1~6个核苷酸重复排列(赵相艳, 2014)。SSR在真核生物基因组中广泛分布(罗文永等, 2003),鱼类基因组中绝大多数SSR位于非编码区,但也有少数SSR位于编码区附近,甚至编码区内(李红梅, 2014)。SSR具有数量多、分布广、易检测且符合孟德尔遗传等特点,目前已被广泛应用于遗传多样性分析、遗传图谱构建、物种及亲缘关系鉴定等研究(罗文永等, 2003; 李红梅, 2014; 赵相艳, 2014)。SSR分子标记的开发对于海水鱼类种质资源保护利用及分子辅助育种等工作的开展具有重要意义。然而,目前已发表军曹鱼SSR标记的数量十分有限,亟需开发筛选更多的多态性标记。
相较于传统的文库法、富集法等SSR开发方法,基于高通量测序数据批量开发SSR标记这一研究策略已在大黄鱼(Pseudosciaena crocea)(李红梅, 2014)、黄鳍棘鲷(Acanthopagrus latus)(吴仁协等, 2019)、红鳍笛鲷(Lutjanus erythropterus)(郭昱嵩等, 2011)等多种鱼类中获得广泛应用。本研究在军曹鱼Illumina HiSeq X Ten及PacBio Sequel测序平台测序获得数据信息的基础上,利用MicroSatellite (MISA)软件从基因组测序结果中查找SSR位点,并对其分布、数量、组成特征等进行分析;利用从中开发的多态性SSR标记用于养殖群体的遗传多样性分析。筛选获得的SSR标记可用于军曹鱼群体遗传结构分析、重要性状QTL定位和良种选育等研究,以期为军曹鱼增养殖业的健康发展提供有效技术支持。
1 材料与方法 1.1 实验材料基因组测序用军曹鱼取自北部湾养殖群体,剪取其背部肌肉,置于95%的乙醇中储存,用于军曹鱼基因组DNA的提取。
军曹鱼样品为北海养殖群体(RC-BH) 30尾、陵水养殖群体(RC-LS)30尾、硇洲养殖群体(RC-NZ) 30尾、徐闻养殖群体(RC-XW) 20尾和三亚养殖群体(RC-SY) 35尾,共145尾。剪取少量尾鳍保存在95%的乙醇中,于−40 ℃储存,用于基因组DNA提取。
1.2 全基因组数据来源军曹鱼基因测序由武汉菲沙基因信息有限公司完成,基因组大小为575.35 Mb,原始数据已上传至NCBI (登录号PRJNA634421)。
1.3 SSR序列搜索及筛选标准利用MISA1.0软件对军曹鱼全基因组序列进行分析,并对基因组单链DNA上SSR序列的数量和种类进行统计。SSR位点的选择及筛选参数设置参照其他鱼类基因组SSR特征分析结果及筛选依据(段永楠等, 2019; 王耀嵘等, 2020; 黄纬杰等, 2022),选取单核苷酸、二核苷酸、三核苷酸、四核苷酸、五核苷酸和六核苷酸重复;通过设定筛选标准为单核苷酸重复单元的重复次数至少10次,二核苷酸重复至少6次,三核苷酸、四核苷酸、五核苷酸和六核苷酸重复次数至少4次,以筛选多态性SSR位点。
1.4 多态性SSR标记的筛选根据上述筛选得到的SSR位点信息,随机选取适合扩增的100个位点设计并合成引物。所有正向引物在5´端添加16 bp的通用Tag,以随机5尾军曹鱼的混合DNA作为模板,优化PCR条件,并排除扩增无条带、条带模糊及多条带等PCR结果不理想的位点。此后,以不同养殖群体的16尾军曹鱼DNA为模板,利用PCR扩增理想的SSR引物进行多重扩增(Tag修饰引物,各标记正向、反向引物)(Schuelke, 2000)。所有引物由北京擎科生物科技有限公司合成。PCR体系为20 μL:金牌Mix 16.45 μL (北京擎科生物科技有限公司),正向引物(10 μmol/L) 0.15 μL,反向引物(10 μmol/L) 1.2 μL,Tag引物(10 μmol/L) 1.2 μL,gDNA 10~50 ng。PCR条件:98 ℃预变性2 min;98 ℃变性10 s,各引物适宜退火温度10 s,72 ℃延伸10 s,共进行35个循环;72 ℃延伸5 min。PCR产物由北京擎科生物科技有限公司进行毛细管荧光电泳检测(ABI3730),并利用GeneMapper 4.1软件进行数据准确位点的分析。基于基因分型数据,挑选其中多态性高的SSR位点进行5个养殖群体的遗传多样性分析。
1.5 数据分析群体遗传多样性的数据统计及分析参照李伟强等(2020)的方法。利用PopGene32软件计算等位基因数(Na)、有效等位基因数(Ne)、Shannon指数(H)、近交系数(Fis)、基因流(Nm)等;利用Cervus 3.0.7软件(Kalinowski et al, 2007)计算微卫星标记观测杂合度(Ho)、期望杂合度(He)和多态信息含量(PIC);利用Genepop4.0.7软件检测哈迪–温伯格(Hardy-Weinberg, HWE)平衡。利用MEGA 6.0软件构建5个军曹鱼养殖群体的系统进化树。
2 结果与分析 2.1 军曹鱼基因组SSR位点类型与数量分析统计军曹鱼基因组中不同碱基重复序列,共发现424 827条SSR重复序列,其中,符合本研究筛选标准的以1~6个碱基为重复单元的SSR位点共计344 820个,占总SSR的81.17%。在这些SSR位点中,单核苷酸重复类型最多(174 146个),占总数的50.50%;其次为二核苷酸重复(104 242个),占总SSR的30.23%;三核苷酸、四核苷酸、五核苷酸和六核苷酸重复各有48 358、12 792、4 403和879个,分别占总SSR的14.02%、3.71%、1.28%和0.25%(表 1)。
|
|
表 1 军曹鱼基因组SSR不同重复单元分布情况 Tab.1 Distribution of different nucleotide repeat types of microsatellite in cobia genome |
由表 2可知,在军曹鱼基因组SSR中,单核苷酸重复序列以A/T重复类别最多,共159 774条,占SSR总数的46.34%;二核苷酸重复序列中,AC/GT为主要重复类别,有75 190条,占SSR总数的21.81%,CG/CG重复类别数量最少(105条),占SSR总数的0.03%;三核苷酸重复序列以AAT/ATT和AGG/CCT居多,分别为13 133和8 775条,各占SSR总数的3.81%和2.54%;四核苷酸重复以AAAT/ATTT出现频率最高,这一重复单元序列共有2 452条,占SSR总数的0.71%;五核苷酸重复序列中AGAGG/CCTCT重复类别最多,有913条,占SSR总数的0.25%;六核苷酸重复序列的重复类别种类多于其他核苷酸,以ACCAGG/CCTGGT为主,仅有92条。
|
|
表 2 军曹鱼SSR重复单元的类型 Tab.2 Type of microsatellite repeat motifs in cobia genome |
军曹鱼基因组SSR核心序列重复次数整体在4~275范围内波动,不同类型SSR核苷酸重复次数的波动范围和数量差异较大。单核苷酸SSR核心序列的重复次数为10~105,其中,重复数为10的SSR最多,有37 572个,占单核苷酸SSR总数的21.58%,20次及以上的重复共计24 812个,在单核苷酸SSR中占14.25%;二核苷酸SSR的重复数为6~167,其中,6次重复的SSR最多,有24 371个,占其总数的23.38%;三核苷酸SSR重复次数为4~275,其中,4次重复最多(29 144个),占SSR总数的60.27%;四核苷酸、五核苷酸、六核苷酸的重复次数分别为4~272、4~28和4~19,均以4次重复最多,分别有7 200、2 290和602个,在同类型的SSR中各自占比56.29%、52.01%和68.49%。
2.4 军曹鱼基因组SSR的长度为筛选具有潜在利用价值的高多态性SSR位点,本研究只对长度≥12 bp的SSR位点进行分析,共筛选出符合上述标准的SSR位点361 684个。由军曹鱼基因组SSR片段长度的分布情况(图 1)可知,片段长度为12~19 bp的SSR位点最多(188 166个),占SSR总数的52.02%;其次是重复单元长度为20~30 bp的SSR位点(70 081个),占SSR总数的19.38%;长度为31~40 bp的SSR位点共有22 809个,占SSR总数的6.31%;长度为41~50 bp的SSR位点有14 551个,占SSR总数的4.02%;片段长度 > 50的SSR数量为66 077个,占SSR总数的18.27%。66 077个,占SSR总数的18.27%。

|
图 1 军曹鱼SSR位点重复单元序列长度分布频率 Fig.1 Frequency of SSR repeat sequence length distribution in cobia |
基于100个候选位点设计的引物在5尾军曹鱼混合DNA的扩增结果中,筛选出条带明亮单一的引物共23对。随后,对筛选出的引物进行荧光标记修饰,并检测其在16尾军曹鱼DNA中的基因分型结果,共从中筛选到10个多态性较高的SSR标记(表 3)。利用这些标记在5个养殖群体145尾军曹鱼中共检测到69个等位基因,其中,最小等位基因数目为5 (RCSSR155),最大等位基因数目为11 (RCSSR50)。观测杂合度(Ho)为0.396~0.799,平均值为0.628;期望杂合度(He)为0.610~0.767,平均值为0.706;平均多态信息含量(PIC)为0.528~0.724,平均值为0.653。10个位点在5个军曹鱼养殖群体中的近交系数(Fis)范围为−0.317~0.270 (表 4)。
|
|
表 3 10个军曹鱼微卫星位点引物基本信息 Tab.3 Basic information of ten identified SSR primers for cobia |
|
|
表 4 军曹鱼10个多态性微卫星位点的遗传学特征 Tab.4 Characterization of 10 polymorphic SSR loci in in five cultured population of cobia |
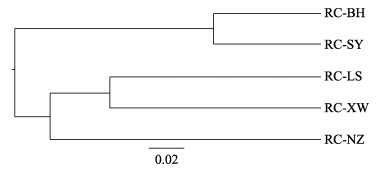
5个军曹鱼养殖群体间的遗传相似度和遗传距离如表 5所示,遗传距离为0.141~0.464,相似度为0.629~0.868。根据Nei´s遗传距离构建的UPGMA系统进化树如图 2所示,RC-LS和RC-XW聚为一支后与RC-NZ聚在一起;RC-BH和RC-SY聚为一支。
|
|
表 5 5个军曹鱼养殖群体的遗传相似度和遗传距离 Tab.5 Genetic identity and genetic distance of five cultured population of cobia |
|
图 2 基于Nei´s遗传距离构建的5个军曹鱼群体的UPGMA系统进化树 Fig.2 UPGMA dendrogram of five population of cobia based on Nei´s genetic distance |
随着SSR重复单元序列碱基数目的增加,军曹鱼基因组SSR不同重复单元的出现频率逐渐下降,与斑点叉尾
军曹鱼基因组SSR中,单核苷酸重复类别主要以A/T类型为主,这与红鳍东方鲀(Takifugu rubripes) (崔建洲等, 2006)、西施舌(王雨吉等, 2021)等物种基因组SSR中的主要单核苷酸重复类型相同。二核苷酸重复类别中,AC/GT类型出现的频率最高,这与黄唇鱼(Bahaba flavolabiata)(赵彦花等, 2019)、大泷六线鱼(Hexagrammos otakii)(沈朕, 2017)、鲤鱼(梁霞等, 2021)等硬骨鱼类的结果一致。此外,在已报道的脊椎动物基因组SSR特征分析结果中,CG/CG这一类型的二核苷酸重复序列的分布水平普遍较低,同样地,军曹鱼基因组SSR中CG/CG类型中在二核苷酸重复类别中的出现次数最少。推测产生上述现象的原因有二,一是少量的CG利于维持DNA热力学稳定性;二是基因组DNA在复制及转录过程中C/G可能突变为A/T (Schorderet et al, 1992)。军曹鱼基因组SSR三核苷酸重复类别中,AAT/ATT重复单元的占比最高。这一优势类型在斑点叉尾
随着核心序列拷贝数的增加,军曹鱼基因组中各重复类型SSR的数目呈递减趋势。这一现象同样存在于斑点叉尾
SSR片段长度大小会影响其多态性的高低,SSR分子标记的多态性越高,其潜在利用价值则越高。当SSR的重复单元长度低于12 bp时,其多态性较低;当SSR重复单元长度大小在12~20 bp区间时,其具有中等多态性水平;当长度 > 20 bp时,其多态性较高(田镇等, 2021)。军曹鱼基因组中,重复序列长度在20 bp以上的SSR位点共有173 518个,占SSR总数的47.98%,这些SSR位点在理论上具有较高的多态性,本研究从中随机选取100个位点进行多态性标记筛选,共获得10个多态性较高的SSR标记,并将其应用于军曹鱼养殖群体遗传多样性分析。
本研究采用李伟强等(2020)使用的相同军曹鱼养殖群体样品,10个SSR位点在5个群体中获得的Na、Ne均低于前期报道中12个SSR位点所获得的数值,但Ho和He则高于或接近前期结果。上述结果表明,利用不同标记对相同养殖群体的遗传多样性评估结果存在一定差异,但各参数的平均值并无显著差异。此外,遗传距离和遗传相似度分析结果显示,各参数数值排序与前期研究相似,即RC-SY与RC-BH之间的遗传距离最小(0.141),而RC-NZ与RC-BH之间的最大(0.464),表明两批次标记对各养殖群体间遗传关系的评价较为一致。基于UPMGA法构建的群体聚类分析结果显示,RC-NZ与RC-LS/RC-XW分支先聚在一起,而后与RC-BH/RC-SY进行聚类;但前期研究结果中,RC-BH和RC-SY聚为一支,RC-LS和RC-XW聚为一支,两支聚为一支后又与RC-NZ聚为一支。由此可见,本研究基于基因组信息查找筛选获得的SSR标记在5个养殖群体中的遗传多样性参数、群体遗传距离等结果均与前期研究相似,进一步验证了军曹鱼养殖群体遗传多样性在多代的人工选育后有所降低这一现状;但群体聚类分析结果的差异也在一定程度上反映了标记数量及类型对遗传多样性评估结果的影响,因此,针对不同情况选取最适数量及类型的标记对于获得准确研究结果至关重要。此外,SSR位点的无效等位基因频率及多态性等因素也会影响聚类分析结果的可靠性。与前期研究相比,本研究筛选得到的SSR标记偏离哈迪–温伯格平衡的情况更为突出,因此,也会造成系统进化树分支的聚类存在差异。最后,鉴于军曹鱼各养殖群体间仍存在较大的分化程度,因此,在后续选育过程中,可利用本研究筛选获得的多态性标记对亲本群体进行评估,优化繁殖配组方式,避免因近亲交配等产生种质退化问题,保障军曹鱼种业的健康发展。
本研究开展的军曹鱼基因组SSR特征分析可为多态性SSR标记的筛选提供重要数据资料,相关研究为鱼类多态性分子标记的开发提供了新思路,筛选获得的SSR标记为鱼类群体遗传多样性分析、种质资源评估及分子辅助育种等提供了有力的评价工具。
BAI C C, LIU S F, ZHUANG Z M. Characteristic analysis of microsatellite DNA in the genome of Gobiidae. Progress in Fishery Sciences, 2016, 37(5): 9-15 [白翠翠, 柳淑芳, 庄志猛. 虾虎鱼科(Gobiidae)基因组微卫星DNA的分布特征. 渔业科学进展, 2016, 37(5): 9-15] |
CHEN G, ZHANG J D, YE N, et al. Introduction of culture technology of cobia, Rachycentron canadum (Ⅰ). Scientific Fish Farming, 2004(1): 10-11 [陈刚, 张健东, 叶宁, 等. 军曹鱼的养殖技术介绍(上). 科学养鱼, 2004(1): 10-11] |
CUI J Z, SHEN X Y, YANG G P, et al. The analysis of simple sequence repeats in Takifugu rubripes genome. Journal of Ocean University of China (Natural Sciences), 2006, 36(2): 249–254, 272 [崔建洲, 申雪艳, 杨官品, 等. 红鳍东方鲀基因组微卫星特征分析. 中国海洋大学学报(自然科学版), 2006, 36(2): 249–254, 272] |
DUAN Y N, LIU Y, HU Y C, et al. Distribution regularity of microsatellites in Scleropages formosus genome. Chinese Agricultural Science Bulletin, 2019, 35(23): 152-158 [段永楠, 刘奕, 胡隐昌, 等. 美丽硬仆骨舌鱼全基因组微卫星分布规律特征. 中国农学通报, 2019, 35(23): 152-158] |
ELLEGREN H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nature Genetics, 2000, 24(4): 400-402 DOI:10.1038/74249 |
GOU X W, OU Y J, LIAO R. Present status on studies of cobia Rachycentron canadum in China. Marine Fisheries, 2007, 29(1): 84-89 [勾效伟, 区又君, 廖锐. 我国军曹鱼研究现状. 海洋渔业, 2007, 29(1): 84-89] |
GUO Y S, WANG Z D, XIE Z Q, et al. Isolation and genetic diversity analysis of microsatellite DNA in Lutjanus erythopterus. Journal of Guangdong Ocean University, 2011, 31(4): 13-17 [郭昱嵩, 王中铎, 谢子强, 等. 红鳍笛鲷微卫星DNA标记的开发与遗传多样性分析. 广东海洋大学学报, 2011, 31(4): 13-17] |
HUANG W J, GUO X Z, ZHANG Z H, et al. Analysis of microsatellite in grass carp (Ctenopharyngodon idella) genome and the application in parentage identification. Journal of Fisheries of China, 2022, 46(2): 1-12 [黄纬杰, 郭向召, 张子豪, 等. 草鱼全基因组微卫星特征分析与亲子鉴定. 水产学报, 2022, 46(2): 1-12] |
KALINOWSKI S T, TAPER M L, MARSHALL T C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology, 2007, 16(5): 1099-1106 DOI:10.1111/j.1365-294X.2007.03089.x |
KUANG J H, CHEN G, MA Q, et al. Embryonic development and morphological characteristics of larvae and juveniles of cobia (Rachycentron canadum). Journal of Fisheries of China, 2021, 45(11): 1814-1824 [邝杰华, 陈刚, 马骞, 等. 军曹鱼的胚胎发育及仔稚鱼形态观察. 水产学报, 2021, 45(11): 1814-1824] |
KUANG J H, CHEN G, MA Q, et al. Histological observation on gonadal differentiation and first annual gonadal development of cobia (Rachycentron canadum). Haiyang Xuebao, 2021, 43(8): 128-138 [邝杰华, 陈刚, 马骞, 等. 军曹鱼(Rachycentron canadum)性腺分化及首周年发育的组织学观察. 海洋学报, 2021, 43(8): 128-138] |
LI H M. New microsatellite satellite markers development based on whole genome sequencing information and its application in population genetics in large yellow croaker. Master´s Thesis of Zhejiang Ocean University, 2014 [李红梅. 基于基因组信息的大黄鱼(Pseudosciaena crocea)微卫星标记开发及应用. 浙江海洋学院硕士研究生学位论文, 2014]
|
LI L D, CHEN B S, FENG J, et al. Analysis and evaluation in nutritive value of Rachycentron canadum (Linnaeus). Journal of Tropical Oceanography, 2002(1): 76-82 [李刘冬, 陈毕生, 冯娟, 等. 军曹鱼营养成分的分析及评价. 热带海洋学报, 2002(1): 76-82] |
LI W Q, CHEN G, MA Q, et al. Genetic diversity in five cultured population of cobia (Rachycentron canadum) using microsatellite markers. Progress in Fishery Sciences, 2020, 41(2): 113-120 [李伟强, 陈刚, 马骞, 等. 利用微卫星标记分析军曹鱼养殖群体的遗传多样性. 渔业科学进展, 2020, 41(2): 113-120] |
LI W, LI J X, JING H F, et al. Development of microsatellite loci for Dabry′s sturgeon (Acipenser dabryanus) using high-throughput sequencing. Chinese Journal of Zoology, 2017, 52(3): 449-457 [李薇, 李久煊, 荆慧芳, 等. 基于高通量测序的达氏鲟微卫星标记筛选. 动物学杂志, 2017, 52(3): 449-457] |
LI X M, WU X B, YANG D G, et al. Development of microsatellite markers for Coreius guichenoti based on transcriptome sequencing. Journal of Hydroecology, 2021, 42(4): 97-103 [李学梅, 吴兴兵, 杨德国, 等. 基于转录组测序的圆口铜鱼微卫星标记筛选. 水生态学杂志, 2021, 42(4): 97-103] |
LIANG X, WANG H Q, MA Y X, et al. Distribution characteristics of microsatellites in the whole genome of Cyprinus carpio, Linnaeus. Journal of Nanjing Normal University (Natural Science), 2021, 44(3): 103-111 [梁霞, 王慧琪, 马宇璇, 等. 鲤鱼(Cyprinus carpio)全基因组微卫星分布特征研究. 南京师大学报(自然科学版), 2021, 44(3): 103-111] |
LIU B, SHAO Y Q, TENG S S, et al. Characterization, development and utilization of EST-derived microsatellites in Sinonovacula constricta. Oceanologia et Limnologia Sinica, 2012, 43(1): 132-137 [刘博, 邵艳卿, 滕爽爽, 等. 缢蛏(Sinonovacula constricta) EST-SSR分布特征及引物开发利用. 海洋与湖沼, 2012, 43(1): 132-137] |
LIU H J, CHEN R F, PENG Y H, et al. Artificial breeding of Rachycentron canadum. Guangxi Sciences, 2012, 19(3): 293-296 [刘海娟, 陈瑞芳, 彭银辉, 等. 军曹鱼人工繁育试验研究. 广西科学, 2012, 19(3): 293-296] |
LUO W Y, HU J, LI X F. The evolution and application of microsatellites. Hereditas(Beijing), 2003, 25(5): 615-619 [罗文永, 胡骏, 李晓方. 微卫星序列及其应用. 遗传, 2003, 25(5): 615-619] |
MA Y C, ZHANG G R, LI M M, et al. Progress in nutritional requirements and feed research of cobia Rachycentron canadum. Acta Hydrobiologica Sinica, 2019, 43(3): 680-692 [麻永财, 张关荣, 李孟孟, 等. 军曹鱼营养需求与饲料研究进展. 水生生物学报, 2019, 43(3): 680-692] |
MAO F F, CHEN G, MA Q, et al. Skeletal deformities in the juveniles of cultured cobia (Rachycentron canadum). Progress in Fishery Sciences, 2022, 43(1): 133-140 [毛非凡, 陈刚, 马骞, 等. 养殖军曹鱼稚鱼骨骼畸形研究. 渔业科学进展, 2022, 43(1): 133-140] |
NEI M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the United States of America, 1973, 70(12): 3321-3323 |
SCHORDERET D F, GARTLER S M. Analysis of CpG suppression in methylated and nonmethylated species. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(3): 957-961 |
SCHUELKE M. An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology, 2000, 18(2): 233-234 |
SHANGGUAN Q, CHEN K C, LIU H Y, et al. Characteristics of microsatellites and genetic structure of wild Channa maculate. South China Fisheries Science, 2020, 16(3): 47-60 [上官清, 陈昆慈, 刘海洋, 等. 斑鳢基因组中微卫星分布特征及野生种群遗传结构分析. 南方水产科学, 2020, 16(3): 47-60] |
SHEN Z. Characterization of novel molecular markers, correlation with growth traits and research of genetic diversity. Master´s Thesis of Shandong University, 2017 [沈朕. 大泷六线鱼分子标记的开发、生长性状的关联性分析及遗传多样性研究. 山东大学硕士研究生学位论文, 2017]
|
SU M Y, YANG W S, TANG R Y, et al. Microsatellite distribution in the whole genome of Ageneiosus marmoratus. Journal of Nanjing Normal University (Engineering and Technology), 2021, 21(2): 65-71 [苏孟园, 杨汶珊, 唐荣叶, 等. 花斑无须鲶(Ageneiosus marmoratus)全基因组微卫星分布特征研究. 南京师范大学学报(工程技术版), 2021, 21(2): 65-71] |
TANG R Y, SU M Y, YANG W S, et al. Analysis of microsatellite distribution characteristics in the channel catfish (Ictalurus punctatus) genome. Progress in Fishery Sciences, 2022, 43(2): 89-97 [唐荣叶, 苏孟园, 杨汶珊, 等. 斑点叉尾  全基因组微卫星分布特征分析. 渔业科学进展, 2022, 43(2): 89-97] 全基因组微卫星分布特征分析. 渔业科学进展, 2022, 43(2): 89-97] |
TIAN Z, CHEN A H, WU Y P, et al. Bioinformatics analysis of microsatellite sites in the RNA-sequencing of Meretrix meretrix. Marine Fisheries, 2021, 43(2): 160-167 [田镇, 陈爱华, 吴杨平, 等. 文蛤转录组中微卫星位点生物信息分析. 海洋渔业, 2021, 43(2): 160-167] |
WANG Y J, MENG X P, YI L F. Isolation of microsatellite markers in transcriptome of Coelomactra antiquata and their application in cryptic species identification. Fisheries Science, 2022, 41(2): 289-295 [王雨吉, 孟学平, 易乐飞. 基于转录组西施舌微卫星标记开发及隐种鉴定. 水产科学, 2022, 41(2): 289-295] |
WANG Y R, YANG W, REN X L, et al. Distribution patterns of microsatellites and development of polymorphic markers from Scatophagus argus genome. Journal of Guangdong Ocean University, 2020, 40(4): 7-14 [王耀嵘, 杨尉, 任席林, 等. 金钱鱼基因组微卫星分布特征分析及多态性标记开发. 广东海洋大学学报, 2020, 40(4): 7-14] |
WANG Z D, CHEN T M, GUO Y S, et al. A genetic analysis of cultured populations of cobia (Rachycentron canadum) with microsatellite markers. Journal of Guangdong Ocean University, 2010, 30(3): 16-21 [王中铎, 陈铁妹, 郭昱嵩, 等. 军曹鱼全人工繁殖群体遗传特征的SSR分析. 广东海洋大学学报, 2010, 30(3): 16-21] |
WANG Z D, SHI P X, SU H N, et al. Analysis on population genetic structure of cobia (Rachycentron canadum) with AFLP markers. Journal of Guangdong Ocean University, 2011, 31(1): 12-17 [王中铎, 史沛鑫, 苏惠娜, 等. 军曹鱼群体遗传结构的AFLP分析. 广东海洋大学学报, 2011, 31(1): 12-17] |
WIERDL M, DOMINSKA M, PETES T D. Microsatellite instability in yeast: Dependence on the length of the microsatellite. Genetics, 1997, 146(3): 769-779 |
WU R X, ZHAI Y, XIAO Y, et al. Microsatellite marker development for Acanthopagrus latus and cross-species amplification in the family Sparidae. Journal of Applied Oceanography, 2019, 38(3): 356-364 [吴仁协, 翟云, 肖瑶, 等. 黄鳍棘鲷微卫星标记开发及其在鲷科鱼类中的跨物种扩增. 应用海洋学学报, 2019, 38(3): 356-364] |
XU J J, ZHENG X, LI J, et al. Distribution characteristics of whole genome microsatellite of Pelteobagrus fulvidraco. Genomics and Applied Biology, 2020, 39(12): 5488-5498 [徐杰杰, 郑翔, 李杰, 等. 黄颡鱼(Pelteobagrus fulvidraco)全基因组微卫星分布特征分析. 基因组学与应用生物学, 2020, 39(12): 5488-5498] |
ZHANG Y D, WEN L T, LUO H L, et al. Genome survey and development of SSR molecular markers for Trachinotus ovatus. Journal of Southern Agriculture, 2020, 51(5): 983-994 [张永德, 文露婷, 罗洪林, 等. 卵形鲳鲹基因组调研及其SSR分子标记的开发应用. 南方农业学报, 2020, 51(5): 983-994] |
ZHAO X Y. Compositional and comparative analysis of microsatellite in organism genomes. Doctoral Dissertation of Hunan University, 2014 [赵相艳. 微卫星序列在生物体基因组中的组成与比较分析. 湖南大学博士研究生学位论文, 2014]
|
ZHAO Y H, OU Y J, WEN J F, et al. Development of SSR markers in Bahaba flavolabiata by transcriptome sequencing. Journal of Southern Agriculture, 2019, 50(9): 2078-2087 [赵彦花, 区又君, 温久福, 等. 基于转录组测序技术的黄唇鱼SSR分子标记筛选. 南方农业学报, 2019, 50(9): 2078-2087] |



