2. 德州市德城区市场监督管理局 山东 德州 253011
2. Decheng Market Supervision Administration, Dezhou 253011, China
中华圆田螺(Cipangopaludina cathayensis)是我国特有的物种(陈元晓等, 2009),适宜生长在富含底泥腐殖质、溶氧充足、天然饵料较丰富的微流水水域中(杨立平, 2007)。稻田养殖田螺优先选取水源丰富、水质清新且水体溶氧达到5 mg/L以上的稻田(廖建云, 2020)。溶解氧是水体环境中最重要的因子之一,对水产动物的生长发育、新陈代谢、繁殖繁育等一系列生命活动都有重要影响(Small et al, 2014)。水中溶解氧低于2 mg/L时,水体处于低氧或缺氧状态(王春枝等, 2014; 王晓雯等, 2016; 徐贺等, 2016)。低氧胁迫会使水生动物生长发育缓慢,抗病能力以及繁殖能力下降,严重时会导致水生动物死亡(徐贺等, 2016)。李丽萍等(2012)在研究溶解氧和体重对无齿蚌耗氧率的影响时发现,椭圆背角无齿蚌(A. woodiana elliptica)和圆背角无齿蚌(A. woodia pacifica)耗氧率均随水中溶解氧降低而降低。
RNA-seq技术是目前筛选机体特异性状的关键基因和研究基因表达调控机制的有效方法(Li et al, 2014)。曹梅等(2021)对低氧胁迫下脊尾白虾(Palaemon carincauda)的转录组分析,推测低氧诱导因子可以激活虾体适应缺氧的一系列生理活动,增强脊尾白虾各类物质代谢,并且能通过抑制线粒体生物合成和活化线粒体自噬来降低线粒体氧耗。刘韬等(2023)基于转录组分析了青田田鱼(Cyprinus carpio var. qingtianensis)对稻田急性溶氧变化的水环境的适应性,为进一步研究青田田鱼在急性溶氧变化环境下的生理调节机制提供了基础数据。
近年来,关于中华圆田螺的研究主要集中在抗肿瘤机制(朱涛等, 2016)、免疫应答(赵劲松等, 2019)、营养价值评价(薛飞等, 2022)和耐抗生素性(王宏伟等, 2017)等方面,而关于中华圆田螺在转录水平上进行的低氧调控方面的研究尚未见报道。本研究基于RNA-seq技术对低氧胁迫下中华圆田螺肝脏组织进行转录组分析,探究低氧胁迫对中华圆田螺的影响,进而为深入研究中华圆田螺应对低氧胁迫的调控机制奠定基础,也为中华圆田螺的健康养殖提供参考。
1 材料与方法 1.1 实验材料实验所用的中华圆田螺均取自广西玉林陆川县养殖基地,其生活水温为(26.5±0.5) ℃,溶氧量(6.9±0.2) mg/L,氨氮含量 < 0.5 mg/L,采用微流水稻田养殖模式。随机选择健康且螺壳无机械损伤的田螺进行实验。选取田螺的壳宽、壳高及体重见表 1。
|
|
表 1 中华圆田螺基础指标 Tab.1 Basic traits of C. cathayensis |
中华圆田螺运至实验室后于40 L塑料桶中暂养1周,温度维持在26 ℃,溶氧量维持在(6.9±0.1) mg/L。将中华圆田螺随机设置为低氧胁迫组和常氧对照组,每组3个重复,每个重复30只。低氧胁迫处理方法:溶解氧在1 h之内从6.9 mg/L下降到2.5 mg/L左右,维持24 h。在低氧培养期间,不投喂饵料,同时利用HQ30D型便携式溶氧仪每1 h测定一次水体中溶氧含量。对照组维持溶氧为(6.9±0.1) mg/L,24 h后与低氧组同时取样。2组各取15只田螺(每个平行组随机取5只田螺),活体解剖,取肝脏组织立即置于液氮中速冻,然后在–80 ℃冰箱中保存备用。
1.3 总RNA的提取分别取低氧组和对照组田螺的肝脏组织(每个平行组随机取5只田螺的肝脏组织并混合),利用Trizol法提取总RNA,通过1%琼脂糖凝胶电泳检测其质量,利用紫外分光光度计检测其纯度及浓度。总RNA的浓度 > 250 ng/μL,OD260 nm/OD280 nm介于1.8~2.2之间,OD260 nm/OD230 nm≥2.0,确保RNA无降解、无污染。
1.4 mRNA文库构建及测序利用试剂盒(Qubit RNA)精确定量RNA,确定构建mRNA文库加入量。根据试剂盒(Hieff NGS™)说明书,用Tris Buffer重悬磁珠对提取的总RNA进行成熟mRNA富集和纯化,按照其建议构建文库。利用Illumina Hiseq™平台进行测序,使用FastQC平台对数据进行可视化质量评估,以确保质量达标。
1.5 转录组序列组装及注释将原始数据使用Trimmomatic去除低质量碱基、含N序列以及reads接头序列,质控后的序列利用Trinity软件组装clean data得到每个转录本中最长序列作为unigene,即参考序列(Grabherr et al, 2011)。将unigene分别与Blast+、NCBI、NR、GO、KEGG、KOG数据库进行比对注释。
1.6 差异表达分析采用DESeq分析差异表达基因(differentially expressed genes, DEGs),筛选阈值为Fold change > 2及Q-value < 0.05,Q-value越小,则基因表达量差异越显著(Anders et al, 2010)。选取DEGs进行GO功能注释和KEGG富集分析。
1.7 实时荧光定量PCR分析选取6个DEGs进行实时荧光定量PCR (RT-qPCR)验证,以此评价RNA-seq技术鉴定差异基因的可靠性。利用Primer 5.0软件设计特异性引物(表 2),并在生工生物工程(上海股份有限公司)完成引物合成。使用反转录试剂盒(TaKaRa)将每组RNA(样本数目及提取方法同1.3)反转录为cDNA,之后以cDNA为模板、β-actin为内参进行RT-qPCR检测。每个样品3次重复,采用2–ΔΔCt方法计算各基因的相对表达量。
|
|
表 2 中华圆田螺肝脏差异表达基因RT-qPCR分析引物 Tab.2 Primers used for the RT-qPCR analysis of differentially expressed genes in the C. cathayensis |
使用Trimmomatic处理Illumina Hiseq™测序得到的原始数据(raw reads),去除低质量与含接头的reads后,利用Trinity软件对所得数据进行组装,得到500 584条转录本(transcript)和232 379条unigenes,转录本与unigenes的N50分别为1 674 bp和1 127 bp。组装数据统计见表 3,unigenes长度分布见图 1。由拼接结果判断出原始数据拼接的质量较高,可用于后续功能注释和DEGs分析。
|
|
表 3 组装结果统计 Tab.3 Statistics of assembly results |

|
图 1 unigenes序列长度分布 Fig.1 Length distribution of unigenes |
将unigenes信息分别在Swiss-Prot、GO、KOG、Nr和KEGG数据库进行序列比对及功能注释。共有232 379条unigenes得到注释,各数据库注释到的unigenes数量及百分比见表 4。
|
|
表 4 Unigenes注释统计 Tab.4 Statistics of unigenes annotated |
与Nr蛋白数据库进行同源性比对后,有26 636条unigenes与已知基因同源,与中华圆田螺相关的注释信息较少(图 2)。注释信息占比最高为海兔(Aplysia california, 6 470),随后依次是霸王莲花青螺(Lottia gigantea, 3 121)、光滑双脐螺(Biomphalaria glabrate, 3 110)、太平洋牡蛎(Crassostrea gigas, 2 976)、舌形贝(Lingula anatine, 994)、双唇海鲶(Octopus bimaculoides, 967)、紫色球海胆(Strongylocentrotus purpuratus, 428)、海葵(Exaiptasia pallida, 370)、囊舌虫(Saccoglossus kowalevskii, 300)和文昌鱼(Branchiostoma floridae, 276)。
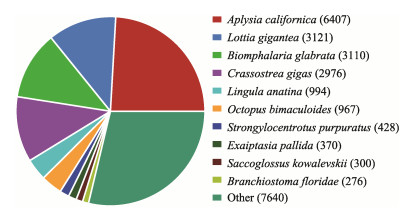
|
图 2 Nr同源物种分布 Fig.2 Homologous species distribution of Nr |
将所有unigenes进行GO统计分析,共有22 907条unigenes得到注释。unigenes在生物学(biological process)、细胞组分(cellular component)和分子功能(molecular function)中均有分布,得到注释的基因数目分别为6 737、8 759、7 411个(图 3)。

|
图 3 Unigenes的GO功能分类 Fig.3 Unigenes GO functional classification |
对所有unigenes进行KOG功能注释,有13 290条unigenes得到注释。其中,信号转导机制(signal transduction mechanisms, 2 543)是最大的类群,占19.13%,其次是转录(transcription, 1 593)和蛋白质翻译后修饰、转换和伴侣(posttranslational modification, protein turnover, chaperones, 1 270)相关的unigenes较多,各占11.99%和9.56%,而与细胞运动(cell motility, 21)相关功能基因占比很少,低于1% (图 4)。
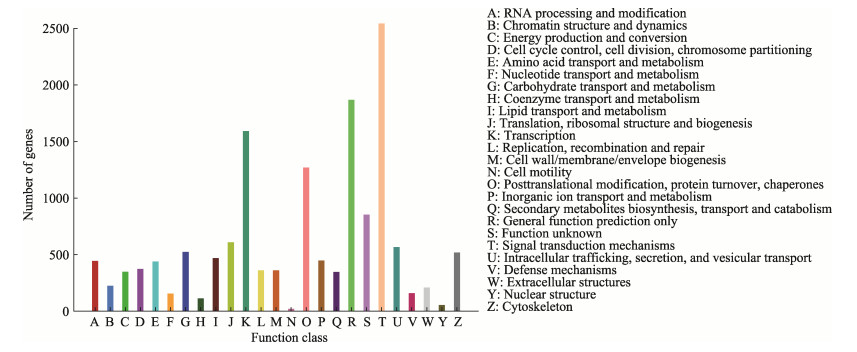
|
图 4 KOG分类柱状图 Fig.4 Column chart of KOG classification |
根据KEGG数据库,进一步对中华圆田螺肝脏unigenes进行注释,共有4 179条unigenes得到注释,分布于新陈代谢(metabolism)、有机系统(organismal systems)、遗传信息处理(genetic information processing)、环境信息处理(environmental information processing)和细胞过程(cellular processes)这5大类通路(图 5)。
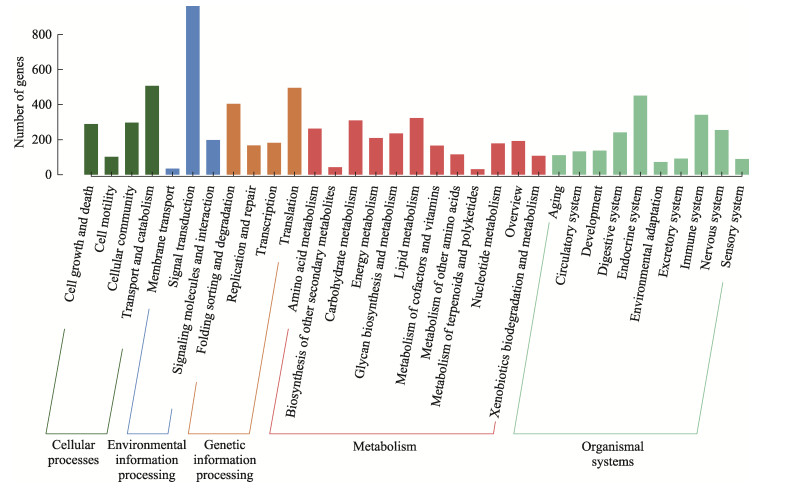
|
图 5 KEGG pathway分类柱状图 Fig.5 KEGG pathway classification histogram |
通过DEseq进行差异表达分析,设置2倍fold change以及Q-value=0.05为筛选标准,综合3次平行重复结果,共得到176个DEGs,包括64条上调基因,112条下调基因(图 6)。
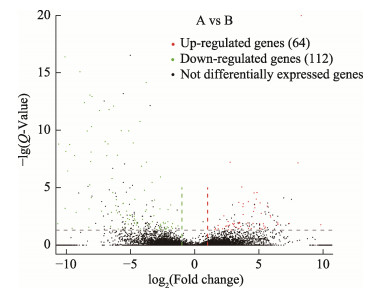
|
图 6 A组和B组差异表达基因统计火山图 Fig.6 Volcano plot of DEGs in A and B groups A:低氧胁迫组;B:对照组 A: Hypoxia group; B: Control group |
与对照组相比,低氧胁迫组中DEGs的GO功能富集分析结果在生物学过程上主要集中于几丁质代谢过程(chitin metabolic process, GO: 0006030)、含氨基葡萄糖的复合代谢过程(glucosamine-containing compound metabolic process, GO: 1901071)和氨基糖代谢途径(amino sugar metabolic process, GO: 0006040)等;在细胞组成上主要集中于胶原三聚体(collagen trimer, GO: 0005581)等;在分子功能上主要集中于几丁质结合(chitin binding, GO: 0008061)、糖衍生物结合(carbohydrate derivative binding, GO: 0097367)、细胞外基质结构组分(extracellular matrix structural constituent, GO: 0005201)和糖衍生物结合(carbohydrate derivative binding, GO: 0097367)等(图 7)。
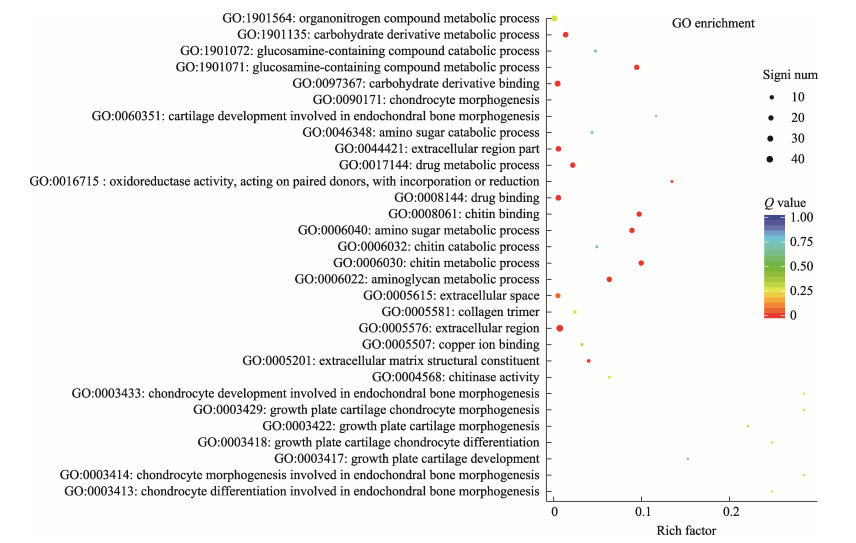
|
图 7 低氧组中华圆田螺肝脏中差异表达基因的GO功能富集图 Fig.7 GO functional enrichment map of DEGs in liver of C. cathayensis under hypoxia group |
对中华圆田螺DEGs进行KEGG pathway富集分析,其注释结果主要集中于环境信息处理(environmental information processing, 3)、遗传信息处理(genetic information processing, 4)、代谢(metabolism, 6)和生物系统(organismal systems, 7)这4大类通路(图 8)。

|
图 8 差异表达基因KEGG分类 Fig.8 KEGG classification map of differentially expressed genes 深色柱体表示所有基因的注释情况,浅色柱体表示差异表达基因的注释情况。 The dark column represents the annotation of unigenes, and the light column represents the annotation of DEGs. |
在这些通路中,信号转导(signal transduction, 3)、折叠排序与降级(folding sorting and degradation, 3)和内分泌系统(endocrine system, 4)的DEGs最多,其次是运输和分解代谢(transport and catabolism, 1)、转录(transcription, 1)、氨基酸代谢(amino acid metabolism, 1)、碳水化合物代谢(carbohydrate metabolism, 2)、脂质代谢(lipid metabolism, 1)、核苷酸代谢(nucleotide metabolism, 1)、衰老(aging, 1)和免疫系统(immune system, 2)。在KEGG pathway中有7个DEGs表达量上调,13个DEGs表达量下调(表 5)。
|
|
表 5 差异表达基因的KEGG富集结果 Tab.5 Enrichment results of KEGG of DEGs |
为验证转录组结果,选取6个DEGs进行RT-qPCR验证。热休克蛋白70B2与热休克蛋白β-6基因表达水平上调;几丁质酶蛋白4 (chitinase-like protein 4)、α-1胶原蛋白(XIV)[collagen alpha-1 chain (XIV)]、α-4胶原蛋白(XIV)、5-磷酸酶蛋白(phosphatase-related protein type 5)基因表达水平下调。结果显示,虽然RT-qPCR定量和RNA-seq表达存在表达量上的偏差,但其变化趋势一致,由此证明了本研究转录组分析结果的可靠性(图 9)。

|
图 9 中华圆田螺肝脏组织差异表达基因RT-qPCR验证 Fig.9 Validation of DEGs in liver tissue of C. cathayensis by RT-qPCR |
高通量测序技术可用于检测基因表达量、揭示转录组复杂性、分析代谢的调控机制等。转录组学连接着生物的基因组遗传信息和蛋白质组功能信息,是基因表达与表型特征的桥梁(Zheng et al, 2011)。梁雪茹等(2022)研究发现,在马氏珠母贝(Pinctada fucata martensii)不同发育阶段的转录组中表达了12个组蛋白H1基因,表明组蛋白H1参与调控马氏珠母贝的生长发育。杨凯等(2020)采用Illumina HiSeq-2500分别对5个不同高温处理组的大菱鲆(Scophthalmus maximus)肾脏组织进行转录组测序,得到68 525条unigenes,初步建立了大菱鲆热应激肾脏转录组数据库。赵晓霞(2011)在基于RNA测序技术的马氏珠母贝珍珠囊转录组及数字基因表达谱分析中发现,被注释的unigenes仅有35.99%。Meyer等(2009)在研究珊瑚虫转录组中发现,只有8.5%的unigenes得到注释,剩余unigenes未能得到注释。本研究中,中华圆田螺转录组组装得到的232 379条unigenes,只有11.46% unigenes得到注释。部分基因未得到注释的原因可能与各类公共基因数据库收录相近物种基因信息较少、注释信息不够全面有关。对中华圆田螺DEGs进行GO功能富集分析,发现DEGs主要与生物学过程中的代谢有关,且集中在氨基糖代谢和几丁质代谢过程。同时,DEGs KEGG pathway分析结果表明,中华圆田螺在低氧胁迫下,通过抑制细胞生长、加速细胞衰老凋亡来应对氧化应激。
3.1 低氧胁迫下热休克蛋白的调控作用热休克蛋白(HSPs)是机体受到外界刺激后而产生的蛋白质,当生物体受到外界刺激时,体内细胞会自动关闭正常的蛋白合成,转而合成一系列的HSPs来快速调整应激过程中细胞的存活性,协助细胞快速适应外界环境的变化,又称为应激蛋白(Zhang et al, 2016)。Yuan等(2010)研究发现,小鼠急速运动后处于低氧状态时,腓肠肌中Hsp70表达量明显上升。HSPs是低氧诱导因子(HIFs)的靶基因,在低氧状态下HIF-1能诱导Hsp70的转录表达(Yeh et al, 2010; Goel et al, 2010)。王婷(2017)通过转录组测序对长牡蛎(Crassostrea gigas)低氧信号通路分子作用机制的研究,分析了HIF与羟化酶PHD的互作关系,发现HIF-α是长牡蛎适应低氧过程的关键基因。曹梅等(2021)基于低氧胁迫下对脊尾白虾进行转录组学分析发现,低氧诱导因子1 (HIF1) 2个亚基HIF1α和HIF1β表达量上调,推测脊尾白虾细胞在低溶氧环境下诱导HIF产生,刺激机体增加血液氧的供应能力。布会敏等(2011)在研究慢性间歇性低压低氧对发育大鼠心肌热休克蛋白表达中发现,大鼠通过增加Hsp27、Hsp70 mRNA的表达来保护心肌细胞免受低氧损伤。由此推测,处于低氧环境胁迫下的中华圆田螺通过提高Hsp70 mRNA的表达量来保护机体肝脏免受低氧损伤,协助机体快速适应环境的改变。本研究中,中华圆田螺肝脏中热休克蛋白Hsp70 B2、Hspβ-6、Hsp9β基因发生了显著上调,由此推测,肝脏中热休克蛋白受低氧胁迫时激发表达特性,从而使中华圆田螺尽可能地避免低氧带来的损伤,提高机体耐低氧能力与存活力。
3.2 低氧胁迫下几丁质的调控作用几丁质又称甲壳质或甲壳素,是以N-乙酰-D-氨基葡萄糖为单体,通过β-1, 4糖苷键连接而成的高分子多聚物(姜竹峪等, 2016)。几丁质的降解与合成是甲壳类动物进行生长发育的重要生理过程,与机体的营养代谢、免疫病害防御等功能密切相关(吕黎等, 2011; Covi et al, 2012)。当水生甲壳类受到外源性刺激时,机体几丁质酶基因表达会发生显著变化,参与生物体的免疫防控(李旭光等, 2017)。王俊等(2017)研究栉孔扇贝(Chlamys farreri)几丁质酶的功能鉴定中推测,几丁质酶通过水解几丁质来保证有机框架的有序性,从而对贝壳外形起到调控作用。宋柳等(2019)通过克隆三疣梭子蟹(Portunus trituberculatus) Group 5的几丁质酶基因PtCht6,分析PtCht6基因作为免疫基因参与机体的病原免疫防御机制。本研究中,通过GO功能富集分析得出低氧胁迫下中华圆田螺DEGs部分集中在几丁质代谢过程中,RT-qPCR验证了几丁质酶蛋白4、胶原α-1蛋白表达基因显著下调,表明低氧胁迫影响了中华圆田螺肝脏中的几丁质、胶原蛋白的合成,可能在软体动物外壳的非生物胁迫响应中起到重要作用。由于目前软体动物的非生物胁迫机理研究较为薄弱(Bueter et al, 2013),这一发现还需进一步验证是否与中华圆田螺耐低氧性状相关,才能进行更深入的研究。
综上所述,本研究为了解中华圆田螺在应对低氧胁迫过程中的生理调节机制,从转录组层面对常氧组和低氧组的中华圆田螺肝脏组织进行研究分析。低氧胁迫24 h后,相对于常氧组,低氧组表达下调的DEGs数目多于表达上调,且多集中于代谢过程,说明中华圆田螺通过抑制更多的代谢通道来应对低氧胁迫下的氧化应激,如降低氨基酸代谢和脂质代谢途径为机体保留能量;而表达上调的HSPs基因说明中华圆田螺通过增强免疫代谢系统来避免机体受到低氧损伤。本研究结果为探究中华圆田螺低氧胁迫响应的分子调控机制提供了基础数据,可为发展中华圆田螺健康养殖技术和进一步研究中华圆田螺低氧胁迫响应的性状提供参考。
ANDERS S, HUBER W. Differential expression analysis for sequence count data. Genome Biology, 2010, 11(10): R106 DOI:10.1186/gb-2010-11-10-r106 |
BU H M, WANG M L, SUN H, et al. Effect of heat shock proteins in cardiac protection of chronic intermittent hypobaric hypoxia in developing rats. Chinese Pharmacological Bulletin, 2011, 27(1): 49-53 [布会敏, 王妹玲, 孙红, 等. 心肌热休克蛋白在发育大鼠慢性间歇性低压低氧中的心脏保护作用. 中国药理学通报, 2011, 27(1): 49-53] |
BUETER C L, SPECHT C A, LEVITZ S M. Innate sensing of chitin and chitosan. PLoS Pathogens, 2013, 9(1): e1003080 DOI:10.1371/journal.ppat.1003080 |
CHEN Y X, CHEN Y J, ZHANG W, et al. Nutrient ingredient analysis and economical value of four species of freshwater snails in Yunnan Province. Sichuan Journal of Anatomy, 2009, 17(2): 28-30 [陈元晓, 陈英杰, 张闻, 等. 云南省4种淡水贝类的营养成分和经济价值. 四川解剖学杂志, 2009, 17(2): 28-30] |
CAO M, WANG X Q, QIN C X, et al. Transcriptome analysis of Palaemon carincauda subjected to hypoxic stress. Progress in Fishery Sciences, 2021, 42(2): 112-123 [曹梅, 王兴强, 秦传新, 等. 脊尾白虾对低氧响应的转录组学分析. 渔业科学进展, 2021, 42(2): 112-123] |
COVI J A, CHANG E S, MYKLES D L. Neuropeptide signaling mechanisms in crustacean and insect molting glands. Invertebrate Reproduction and Development, 2012, 56(1): 33-49 DOI:10.1080/07924259.2011.588009 |
GRABHERR M G, HAAS B J, YASSOUR M, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nature Biotechnology, 2011, 29(7): 644-652 DOI:10.1038/nbt.1883 |
GOEL G, GUO M, DING J, et al. Combined effect of tumor necrosis factor (TNF)-alpha and heat shock protein (HSP)-70 in reducing apoptotic injury in hypoxia: A cell culture study. Neuroscience Letters, 2010, 483(3): 162-166 DOI:10.1016/j.neulet.2010.07.069 |
JIANG Z Y, CHEN Y, WEI J X, et al. Recent advances in research on chitinase. Journal of Shenyang Pharmaceutical University, 2016, 33(5): 414-418 [姜竹峪, 陈羽, 魏锦兴, 等. 几丁质酶的研究进展. 沈阳药科大学学报, 2016, 33(5): 414-418 DOI:10.14066/j.cnki.cn21-1349/r.2016.05.015] |
LIAO J Y. Ecological cultivation technology of snail in paddy field. Rural Technology, 2020(14): 113-114 [廖建云. 稻田生态养殖田螺技术. 乡村科技, 2020(14): 113-114 DOI:10.19345/j.cnki.1674-7909.2020.14.057] |
LI L P, CAI L, TU Z Y, et al. Effects of dissolved oxygen and body weight on the oxygen consumption rate of Anodanta woodiana. Journal of Hydroecology, 2012, 33(5): 61-65 [李丽萍, 蔡露, 涂志英, 等. 溶解氧和体重对背角无齿蚌耗氧率的影响. 水生态学杂志, 2012, 33(5): 61-65] |
LI E, LI C. Use of RNA-seq in aquaculture research. Poultry Fisheries and Wildlife Sciences, 2014, 2: 2 |
LIU T, QI M, GAO Y, et al. Transcriptome analysis of brain tissue of juvenile Qingtian paddy field carp (Cyprinus carpio var. qingtianensis) during acute changes in dissolved oxygen. Journal of Shanghai Ocean University, 2023, 32(1): 79-88 [刘韬, 齐明, 高阳, 等. 青田田鱼幼鱼脑组织在急性低氧胁迫、复氧恢复的转录组分析. 上海海洋大学学报, 2023, 32(1): 79-88] |
LIANG X R, CAO Y F, YANG S, et al. Identification and functional differentiation analysis of histone H1 gene family in Pinctada fucata martensii. Genomics and Applied Biology, 2022, 41(3): 472-481 [梁雪茹, 曹艳飞, 杨帅, 等. 马氏珠母贝组蛋白H1基因家族的鉴定及功能分析. 基因组学与应用生物学, 2022, 41(3): 472-481] |
LÜ L, NING Q J. Research advances on structures and functions of crustacean chitinase genes. Progress in Physiological Sciences, 2011, 42(6): 457-459 [吕黎, 宁黔冀. 甲壳动物几丁质酶基因结构与功能的研究进展. 生理科学进展, 2011, 42(6): 457-459] |
LI X G, ZHOU G, ZHOU J, et al. Research progresses in function and expression regulation of chitinase gene family of aquatic crustaceans. Journal of Aquaculture, 2017, 38(4): 26-30 [李旭光, 周刚, 周军, 等. 水生甲壳类几丁质酶类基因家族功能与表达调控的研究进展. 水产养殖, 2017, 38(4): 26-30] |
MEYER E, AGLYAMOVA G V, SHI W, et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics, 2009, 10(1): 219 |
SMALL K, KOPF R K, WATTS R J, et al. Hypoxia, blackwater and fish kills: Experimental lethal oxygen thresholds in juvenile predatory lowland river fishes. PLoS One, 2014, 9(4): e94524 |
SONG L, LÜ J J, WANG L, et al. Cloning of chitinase gene (PtCht6) in Portunus trituberculatus and its functional analysis in immunity. Oceanologia et Limnologia Sinica, 2019, 50(5): 1080-1090 [宋柳, 吕建建, 王磊, 等. 三疣梭子蟹几丁质酶基因(PtCht6)的克隆及其在免疫中的功能分析. 海洋与湖沼, 2019, 50(5): 1080-1090] |
WANG C Z, LI Z, LIANG H W, et al. The effects of hypoxia stress on mitochondrial ATPase activity and the expression of F1-δ in Hypophthalmichthys molitrix. Journal of Fishery Sciences of China, 2014, 21(3): 454-463 [王春枝, 李忠, 梁宏伟, 等. 低氧胁迫对鲢线粒体ATP酶活性及F1-δ基因表达的影响. 中国水产科学, 2014, 21(3): 454-463] |
WANG X W, ZHU H, HU H X, et al. Effects of hypoxia on physiological status of siberian sturgeon Acipenser baeri juveniles. Fisheries Science, 2016, 35(5): 459-465 [王晓雯, 朱华, 胡红霞, 等. 低氧胁迫对西伯利亚鲟幼鱼生理状态的影响. 水产科学, 2016, 35(5): 459-465] |
WANG H W, LI L F, ZHAO W B, et al. The effect of roxithromycin on the activity of ERND in Cipangopaludina chinensis. Journal of Shanghai Ocean University, 2017, 26(3): 400-405 [王宏伟, 李林锋, 赵文博, 等. 罗红霉素对中国圆田螺ERND酶活性的影响. 上海海洋大学学报, 2017, 26(3): 400-405] |
WANG T. The molecular mechanism of hypoxia signal pathway in the Pacific oyster (Crassostrea gigas). Doctoral Dissertation of University of Chinese Academy of Seiences, 2017 [王婷. 长牡蛎低氧信号通路分子作用机制研究. 中国科学院大学博士研究生学位论文, 2017]
|
WANG J, GAO J, XIE J, et al. Cloning and mineralization-related functions of the chitinase gene in Chlamys farreri. Journal of Fisheries of China, 2017, 41(2): 189-197 [王俊, 高静, 谢军, 等. 栉孔扇贝几丁质酶基因的克隆及功能鉴定. 水产学报, 2017, 41(2): 189-197] |
XU H, CHEN X M, WANG G Q, et al. Research progress of hypoxia on aquaculture. Feed Industry, 2016, 37(2): 33-37 [徐贺, 陈秀梅, 王桂芹, 等. 低氧胁迫在水产养殖中的研究进展. 饲料工业, 2016, 37(2): 33-37] |
XUE F, HUANG K, SU Z J, et al. Analysis and evaluation of amino acids, fatty acids and safety of heavy metals in Cipangopaludina cathayensis. Progress in Fishery Sciences, 2022, 43(1): 180-187 [薛飞, 黄凯, 宿志健, 等. 中华圆田螺氨基酸、脂肪酸营养价值与重金属安全性评价. 渔业科学进展, 2022, 43(1): 180-187] |
YANG L P. Cultivation techniques of Cipangopaludina chinensis. Inland Fisheries, 2007, 32(1): 40 [杨立平. 中华圆田螺养殖技术. 内陆水产, 2007, 32(1): 40] |
YANG K, HUANG Z H, MA A J, et al. Transcriptome study of kidney of turbot under high-temperature stress. Progress in Fishery Sciences, 2020, 41(1): 86-95 [杨凯, 黄智慧, 马爱军, 等. 高温胁迫条件下大菱鲆肾脏转录组研究. 渔业科学进展, 2020, 41(1): 86-95] |
YUAN Z Q, ZHANG Y, LI X L, et al. HSP70 protects intestinal epithelial cells from hypoxia/reoxygenation injury via a mechanism that involves the mitochondrial pathways. European Journal of Pharmacology, 2010, 643(2/3): 282-288 |
YEH C H, HSU S P, YANG C C, et al. Hypoxic preconditioning reinforces HIF-alpha-dependent HSP70 signaling to reduce ischemic renal failure-induced renal tubular apoptosis and autophagy. Life Sciences, 2010, 86(3/4): 115-123 |
ZHU T, XU L F, LIU X Y, et al. Study on the inhibitive effect of polysaccharide in snail on tumor cells in vitro. Journal of Yunnan University of Traditional Chinese Medicine, 2016, 39(1): 13-16 [朱涛, 许礼发, 刘小燕, 等. 田螺多糖抑制肿瘤细胞增殖生物活性的实验研究. 云南中医学院学报, 2016, 39(1): 13-16] |
ZHAO J S, TONG C C, ZHOU J W, et al. A comparative study of phenoloxidase activity between Cipangopaludina cathayensis and Oncomelania hupensis. Chinese Journal of Schistosomiasis Control, 2019, 31(2): 169–170, 181 [赵劲松, 童成成, 周佳维, 等. 湖北钉螺与中华圆田螺体内酚氧化酶活力比较. 中国血吸虫病防治杂志, 2019, 31(2): 169–170, 181] |
ZHENG W L, WANG Z Y, COLLINS J E, et al. Comparative transcriptome analyses indicate molecular homology of zebrafish swimbladder and mammalian lung. PLoS One, 2011, 6(8): e24019 |
ZHAO X X. Studies on transcriptome and digital gene expression of pearl sac in Pinctada martensii by RNA sequencing. Master´s Thesis of Guangdong Ocean University, 2011 [赵晓霞. 基于RNA测序技术的马氏珠母贝珍珠囊转录组及数字基因表达谱分析. 广东海洋大学硕士研究生学位论文, 2011]
|
ZHANG Q, SHANG M, ZHANG M, et al. Microvesicles derived from hypoxia/reoxygenation-treated human umbilical vein endothelial cells promote apoptosis and oxidative stress in H9c2 cardiomyocytes. BMC Cell Biology, 2016, 17(1): 25 |



