2. 上海海洋大学水产科学国家级实验教学示范中心 上海 201306;
3. 天津农学院水产学院 天津 300384
2. National Demonstration Center for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai 201306, China;
3. College of Fisheries, Tianjin Agricultural University, Tianjin 300384, China
杀鲑气单胞菌(Aeromonas salmonicida)属气单胞菌科(Aeromonadaceae)、气单胞菌属(Aeromonas),为兼性好氧或需氧革兰氏阴性菌(Najimi et al, 2008),主要分为5个亚种:杀鲑亚种(A. salmonicida subsp. salmonicida)、史氏亚种(A. salmonicida subsp. smithia)、无色亚种(A. salmonicida subsp. achromogenes)、杀日本鲑亚种(A. salmonicida subsp. masoucida)和溶果胶亚种(A. salmonicida subsp. pectinolytica) (Pavan et al, 2000)。杀鲑气单胞菌在国内已报道可以感染多种养殖鱼类并引起疖疮病(张晓君等, 2006; 李绍戊等, 2015; 周冬仁等, 2015; 王晓冉等, 2017)。传统的杀鲑气单胞菌鉴定方法主要根据生理生化特征、菌株形态和培养特性等特征,这些方法繁琐耗时且易与其他病原菌发生交叉反应(Lee et al, 2002; Keeling et al, 2013)。16S rRNA基因扩增已用于杀鲑气单胞菌的鉴定(del Cerro et al, 2002; Hiney et al, 1998; Miyata et al, 1996),其他常规PCR (Byers et al, 2002)、多重PCR (Altinok et al, 2008)、荧光定量PCR (Fernández-Álvarez et al, 2016)、环介导等温扩增技术(LAMP) (Kulkarni et al, 2009)和免疫学技术(Adams et al, 1990)也有广泛报道,但这些方法只能鉴定属或种,无法鉴定到亚种。目前,仅有2篇关于杀鲑气单胞菌杀鲑亚种快速检测方法的报道,但根据最新的基因组信息数据,这些方法均具有一定的局限性。Miyata等(1996)利用PCR方法建立了杀鲑气单胞菌杀鲑亚种的检测方法,但加拿大分离的杀鲑亚种中缺少该方法的目标基因;Fernández-Álvarez等(2016)建立了SYBR GreenⅠ荧光定量PCR方法检测杀鲑气单胞菌杀鲑亚种,灵敏度可达到21 fg,但在部分杀日本鲑亚种基因组中也存在检测的目标基因。
基于上述原因,为建立杀鲑气单胞菌亚种的检测方法,本研究以实验室前期分离到的杀鲑气单胞菌杀鲑亚种和杀日本鲑亚种为研究对象,通过分析已公布的杀鲑气单胞菌基因组序列信息,选取phoB基因和LOC111476736基因设计特异性引物,建立杀鲑气单胞菌杀鲑亚种和杀日本鲑亚种的PCR检测方法,为杀鲑气单胞菌病原的快速检测和亚种区分提供技术支持。
1 材料与方法 1.1 菌株及培养条件本实验用菌株见表 1,其中,杀鲑气单胞菌、鳗鱼气单胞菌(Aeromonas encheleia)在TSA培养基20 ℃培养,大肠杆菌(Escherichia coli)在LB培养基37 ℃培养,杀鱼爱德华氏菌(Edwardsiella piscicida)、嗜水气单胞菌(Aeromonas hydrophila)、美人鱼发光杆菌(Photobacterium damselae)、鳗弧菌(Vibrio anguillarum)、哈氏弧菌(Vibrio harveyi)、副乳房链球菌(Streptococcus parauberis)、海豚链球菌(Streptococcus iniae)、停乳链球菌(Streptococcus dysgalactiae)和枯草芽孢杆菌(Bacillus subtilis)等其他菌株在TSA培养基中28 ℃培养。杀鲑气单胞菌标准菌株购自美国菌种保藏中心(ATCC),其他菌株由本实验室临床分离并鉴定。
|
|
表 1 本研究使用菌株 Tab.1 Bacterial strains used in this study |
本实验使用细菌基因组DNA提取试剂盒(天根, 中国)按照说明书步骤提取细菌基因组DNA。
1.3 特异性引物设计和筛选反应条件对GenBank中已公布的48株确定亚种的杀鲑气单胞菌基因组信息进行比较基因组学分析,筛选杀鲑气单胞菌亚种的特异性基因,并将特异性基因的序列与其他病原进行比较分析。根据分析结果,最终选择杀鲑气单胞菌杀鲑亚种phoB基因和杀日本鲑亚种LOC111476736基因作为目标基因。利用Primer- BLAST设计特异性引物,并通过BLAST筛选,排除引物与其他微生物和大菱鲆(Scophthalmus maximus)、大西洋鲑(Salmo salar)、虹鳟(Oncorhynchus mykiss)和褐鳟(Salmo trutta)等杀鲑气单胞菌敏感宿主基因组的非特异性结合,选取最适引物用于后期检测(表 2)。按照KOD酶(TOYOBO, 上海)说明书配制25 μL PCR体系:2.5 µL 10×PCR buffer、0.75 μL 10 µmol/L正/反向引物、1 μL 2 mmol/L dNTPs、1.5 μL 25 mmol/L MgSO4、0.5 μL 1 U/µL KOD酶、1 μL DNA模板、17 μL ddH2O,空白对照以ddH2O作为模板。引物A. salmonicida F1/R1和A. masoucida F2/R2的PCR条件:94 ℃预变性5 min;98 ℃变性10 s,分别采用52、54、56、58、60、62和64 ℃退火30 s,68 ℃延伸15 s,35个循环;68 ℃充分延伸7 min。扩增产物用1.5%琼脂糖凝胶电泳检测后测序验证。
|
|
表 2 本研究中所用引物 Tab.2 Primers used in this study |
为优化杀鲑气单胞菌杀鲑亚种和杀日本鲑亚种的PCR扩增条件,分别对反应体系中的dNTPs浓度、引物浓度、MgSO4浓度和酶浓度进行优化,优化条件见表 3。PCR产物经1.5%琼脂糖凝胶电泳后比较产物的亮度差异。
|
|
表 3 PCR体系的优化(25 µL) Tab.3 Optimization of PCR reaction system (25 µL) |
以平板计数法测定杀鲑气单胞菌杀鲑亚种ATCC33658和杀日本鲑亚种ATCC27013的菌液浓度,用PBS将ATCC33658菌液梯度稀释至1.28×105~ 1.28×100 CFU/μL,将ATCC27013菌液梯度稀释至1.19×105~1.19×100 CFU/μL,分别取1 μL稀释的菌液用于PCR扩增并进行电泳检测。
使用NanoDrop 2000测定杀鲑气单胞菌的基因组DNA浓度,用ddH2O将基因组梯度稀释至1.76×103~1.76×100 fg/μL (ATCC33658)和2.72×103~ 2.72×100 fg/μL (ATCC27013)。每个稀释梯度的DNA溶液分别取1 μL作为模板用于PCR扩增检测。
1.6 PCR特异性检测本实验所用细菌见表 1,用PBS将过夜培养的细菌稀释至约1×106 CFU/mL,95 ℃加热5 min,取1 μL作为模板,以同样浓度的杀鲑气单胞菌杀鲑亚种ATCC33658和杀日本鲑亚种ATCC27013菌体为阳性对照,以ddH2O为阴性对照,利用引物A. salmonicida F1/R1、A. masoucida F2/R2对23株菌进行PCR扩增,并进行电泳检测,确定PCR检测方法的特异性。
1.7 鱼体中的病原检测大菱鲆购于莱阳某养殖场,体重为50~60 g,实验前养殖于100 L循环海水养殖系统中,水温为18 ℃。感染实验前随机取5尾鱼解剖,取肝脏、脾脏和肾脏匀浆涂布于TSA培养基,确定实验鱼未携带病原。将实验鱼随机分为2组,每组10尾。利用PBS缓冲液将培养好的杀鲑气单胞菌杀鲑亚种ASS20200608XZ11L和杀日本鲑亚种ASM20160705RZ6S菌液浓度调整至1×106 CFU/mL。采用背部肌肉注射方法,每尾大菱鲆注射0.1 mL,对照组注射0.1 mL的PBS缓冲液,饲养于相同条件下。取50 mg发病鱼组织加入PBS研磨,95 ℃加热10 min,取1 µL作为模板,ASS20200608XZ11L和ASM20160705RZ6S菌体DNA为阳性对照。按照上述反应体系和程序进行PCR。
2 结果 2.1 PCR检测的最适反应条件利用引物A. salmonicida F1/R1、A. masoucida F2/R2对杀鲑气单胞菌杀鲑亚种ATCC33658和杀鲑气单胞菌杀日本鲑亚种ATCC27013进行PCR,测序结果显示,A. salmonicida F1/R1引物扩增的片段为522 bp,A. masoucida F2/R2引物扩增的片段为515 bp,与预测结果一致,且经比对后与GenBank中phoB、LOC111476736基因的序列完全一致。以引物A. salmonicida F1/R1和A. masoucida F2/R2进行PCR,退火温度为52~64 ℃时均能扩增出特异性条带(图 1),亮度无明显差异且无杂带。为保证特异性,2对引物均以64 ℃作为退火温度。
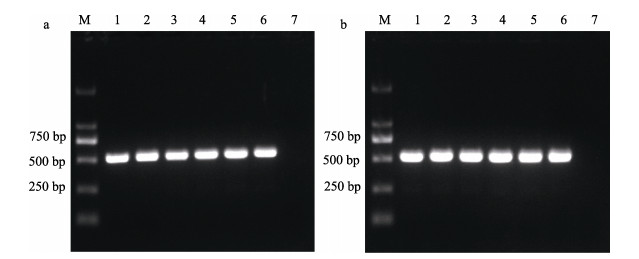
|
图 1 引物A. salmonicida F1/R1 (a)和A. masoucida F2/R2 (b)退火温度筛选 Fig.1 Screening annealing temperature of primers A. salmonicida F1/R1 (a) and A. masoucida F2/R2 (b) M:DL 2000 marker;1~6:退火温度为52、54、56、58、60、62、64 ℃;7:空白对照。 M: DL 2000 marker; 1~6: Annealing temperature of 52, 54, 56, 58, 60, 62, and 64 ℃; 7: Negative control. |
为提高灵敏度,对25 µL PCR体系进行优化,结果表明,以A. salmonicida F1/R1为引物的杀鲑气单胞菌杀鲑亚种PCR检测体系中,2 mmol/L dNTPs最适添加量为2.0、3.0、4.0 µL (图 2a),25 mmol/L MgSO4最适添加量为1.5、2.0、2.5 µL (图 2b),10 µmol/L引物最适添加量为1.5 µL (图 2c),1 U/µL酶最适添加量为0.50、0.75 µL(图 2d)。因此,后期引物A. salmonicida F1/R1采用的反应体系为2.5 µL 10× PCR buffer、2 µL 2 mmol/L dNTPs、1.5 µL 25 mmol/L MgSO4,1.5 µL 10 µmol/L正/反向引物,0.5 µL 1 U/µL酶、15.5 µL ddH2O、1 µL模板。反应程序为94 ℃预变性5 min;98 ℃变性10 s、64 ℃退火30 s、68 ℃延伸15 s,35个循环;68 ℃充分延伸7 min。
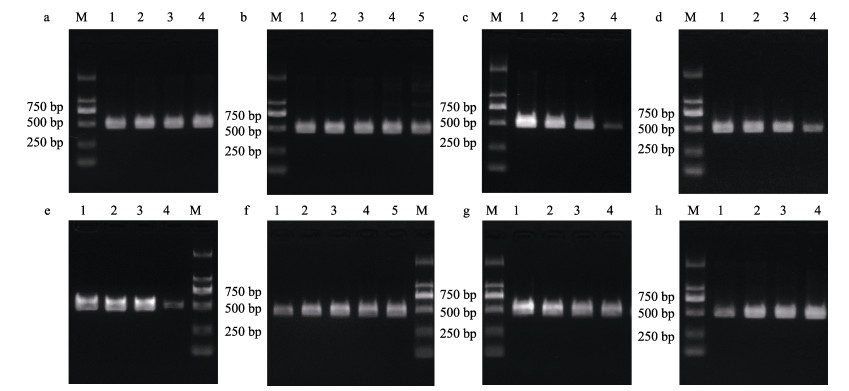
|
图 2 引物A. salmonicida F1/R1 (a、b、c、d)和A. masoucida F2/R2 (e、f、g、h) PCR体系优化 Fig.2 PCR system optimization of primers A. salmonicida F1/R1 (a, b, c, d) and A. masoucida F2/R2 (e, f, g, h) a, e: 2 mmol/L dNTPs, 1~4: 1, 2, 3, 4 µL; b, f: 25 mmol/L MgSO4, 1~5: 1.0, 1.5, 2.0, 2.5, 3.0 µL; c, g: 10 µmol /L primer, 1~4: 1.50, 1.00, 0.75, 0.50 µL; d, h: 1 U/µL KOD, 1~4: 0.25, 0.50, 0.75, 1.00 µL. M: DL 2000 marker. |
以A. masoucida F2/R2为引物的杀鲑气单胞菌杀日本鲑亚种PCR检测体系中,2 mmol/L dNTPs最适添加量为1.0、2.0、3.0 µL (图 2e);25 mmol/L MgSO4最适添加量为1.5、2.0、2.5、3.0 µL (图 2f);10 µmol/L引物的添加量为0.75、1.00、1.50 µL (图 2g);1 U/µL酶最适添加量为0.50、0.75、1.00 µL (图 2h)。因此,后期引物A. masoucida F2/R2采用的反应体系为2.5 µL 10×PCR buffer、1 µL 2 mmol/L dNTPs、1.5 µL 25 mmol/L MgSO4、0.75 µL 10 µmol/L正/反向引物、0.5 µL 1 U/µL酶、17 µL ddH2O、1 µL模板。反应程序为94 ℃预变性5 min;98 ℃变性10 s、64 ℃退火30 s、68 ℃延伸15 s,35个循环;68 ℃充分延伸7 min。
2.3 PCR检测灵敏度以杀鲑气单胞菌杀鲑亚种ATCC33658梯度稀释的菌液为模板,菌液浓度为1.28×105~1.28×101 CFU/μL时均可扩增出目的条带,但当菌液浓度为1.28 CFU/μL时未扩增到目的条带,因此,本方法针对杀鲑气单胞菌杀鲑亚种的检测灵敏度为12.8 CFU/反应(图 3a)。以DNA为模板,当浓度为1.76 fg时未扩增到目的条带,其他浓度均可检测到条带。因此,A. salmonicida F1/R1引物的PCR检测方法检出下限为17.6 fg/反应(图 3b)。
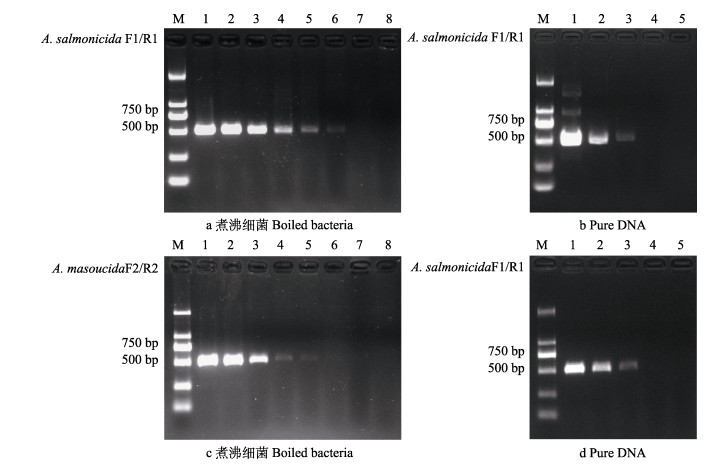
|
图 3 以细菌(a、c)和DNA(b、d)为模板进行PCR灵敏度检测 Fig.3 PCR sensitivity detection using bacteria (a, c) and DNA (b, d) as templates a:1~7:使用引物A. salmonicida F1/R1,每反应以1.28×105、1.28×104、1.28×103、1.28×102、0.256×102、1.28×101和1.28×100 CFU的ATCC33658菌体为模板进行PCR;8:空白对照。b:1~4:使用引物A. salmonicida F1/R1,每反应以1.76×103、1.76×102、1.76×101、1.76×100 fg的ATCC33658基因组DNA为模板进行PCR;5:空白对照。c:1~7:使用引物A. masoucidaF2/R2,每反应以1.19×105、1.19×104、1.19×103、1.19×102、0.238×102、1.19×101和1.19×100 CFU煮沸细菌ATCC27013为模板进行PCR;8:空白对照。d:1~4:使用引物A. masoucida F2/R2,每反应以2.72×103、2.72×102、2.72× 101、2.72×100 fg的ATCC27013基因组DNA为模板进行PCR;5:空白对照。M: DL2000 marker。 a: 1~7: Using primers A. salmonicida F1/R1 and 1.28×105, 1.28×104, 1.28×103, 1.28×102, 0.256×102, 1.28×101 and 1.28×100 CFU boiled bacteria ATCC33658 as template for each PCR; 8: Negative control. b: 1~4: Using primer A. salmonicida F1/R1 and 1.76×103, 1.76×102, 17.6, and 1.76×100 fg of ATCC33658 genomic DNA as the template for each PCR; 5: Negative control. c: Using primers A. masoucida F2/R2 and 1.19×105, 1.19×104, 1.19×103, 1.19×102, 0.238×102, 1.19×101 and 1.19×100 CFU boiled bacteria ATCC27013 as template for each PCR; 8: Negative control. d: 1~4: Using primers A. masoucida F2/R2 and 2.72×103, 2.72×102, 2.72×101 and 2.72×100 fg of ATCC27013 genomic DNA as templates for each PCR; 5: Negative control. M: DL2000 marker. |
以杀鲑气单胞菌杀日本鲑亚种ATCC27013梯度稀释的菌液为模板,菌液浓度为1.19×105~2.38× 101 CFU/μL时均可扩增出目的条带,但当菌液浓度低至1.19×101 CFU/μL时未扩增出目的条带,因此,本方法针对杀鲑气单胞菌杀日本鲑亚种的检测灵敏度为23.8 CFU/反应(图 3c)。以DNA为模板,当浓度为2.72 fg时未扩增到目的条带,其他浓度均可检测到条带。因此,A. masoucida F2/R2引物的PCR检测检出下限为27.2 fg/反应(图 3d)。
2.4 PCR特异性检测选取表 1中的23株菌,包括4株杀鲑气单胞菌杀日本鲑亚种、3株杀鲑气单胞菌杀鲑亚种和14种其他菌属细菌,检测引物A. salmonicida F1/R1、A. masoucida F2/R2的特异性,以A. salmonicida F1/R1为引物进行PCR时,仅在杀鲑气单胞菌杀鲑亚种中扩增出特异性目的条带(图 4a)。以A. masoucida F2/R2为引物进行PCR时,仅杀鲑气单胞菌杀日本鲑亚种扩增出特异性目的条带(图 4b),2对引物从其他细菌的DNA中未扩增出预期DNA条带。因此,本研究设计的2对引物能够特异性地检测杀鲑气单胞菌杀鲑亚种和杀日本鲑亚种。
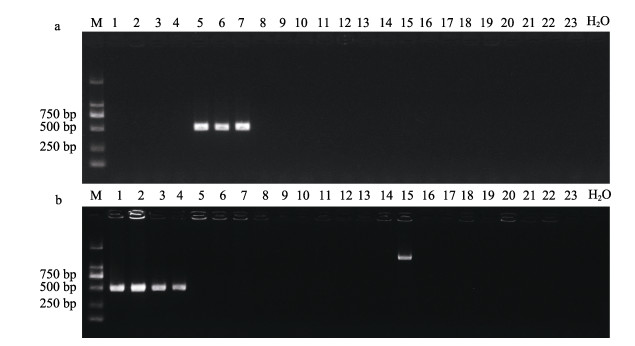
|
图 4 PCR检测引物A. salmonicida F1/R1 (a)和A. masoucida F2/R2 (b)的特异性 Fig.4 Specificity of PCR detection with primers A. salmonicida F1/R1 (a) or A. masoucida F2/R2 (b) M:DL 2000 marker;1~4:杀鲑气单胞菌杀日本鲑亚种;5~7:杀鲑气单胞菌杀鲑亚种;8:杀鲑气单胞菌无色亚种;9:杀鲑气单胞菌史氏亚种;10:大肠杆菌;11:嗜水气单胞菌;12:鳗弧菌;13~14:哈氏弧菌;15:鳗鱼气单胞菌;16:美人鱼发光杆菌;17:美人鱼发光杆菌;18:杀鲑弧菌;19:海豚链球菌;20:副乳房链球菌;21:停乳链球菌;22:杀鱼爱德华氏菌;23:芽孢杆菌;24:空白对照。 M: DL 2000 marker; 1~4: A. salmonicida subsp. masoucida; 5~7: A. salmonicida subsp. salmonicida; 8: A. salmonicida subsp. achromogenes; 9: A. salmonicida subsp. smithia; 10: E. coli; 11: A. hydrophila; 12: V. anguillarum; 13~14: V. harveyi; 15: A. encheleia; 16: P. damselae; 17: P. damselae; 18: V. salmonicida; 19: S. iniae; 20: S. parauberis; 21: S. dysgalactiae; 22: E. piscicida; 23: Bacillus sp.; 24: Blank control. |
以杀鲑气单胞菌杀鲑亚种ASS20200608XZ11L和杀鲑气单胞菌杀日本鲑亚种ASM20160705RZ6S肌肉注射感染大菱鲆,7 d内大菱鲆全部死亡。组织检测结果显示,A. salmonicida F1/R1和A. masoucida F2/R2均能检测到病鱼组织中的杀鲑气单胞菌杀鲑亚种或杀日本鲑亚种,而在健康鱼组织中未扩增出预期条带(图 5),表明引物A. salmonicida F1/R1和A. masoucida F2/R2可以从发病的鱼中检测到杀鲑气单胞菌病原。
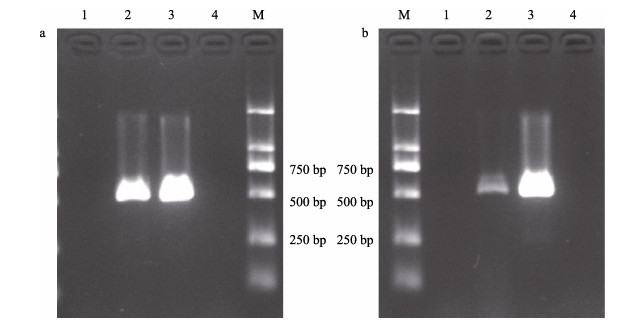
|
图 5 以发病大菱鲆组织为模板进行PCR Fig.5 PCR with infected turbot tissue as template a:引物为A. salmonicida F1/R1进行的PCR;b:引物为A. masoucida F2/R2进行的PCR;1:健康鱼组织液;2:病死鱼组织液;3:阳性对照;4:空白对照;M:DL 2000 marker。 a: PCR with primer A. salmonicida F1/R1; b: PCR with primer A. masoucida F2/R2; 1: Healthy fish tissue fluid; 2: Infected fish tissue fluid; 3: Positive control; 4: Blank control; M: DL 2000 marker. |
杀鲑气单胞菌是水产养殖过程中最为常见的病原菌之一,最早在欧洲的鲑鳟类发现,严重影响了鲑鳟类的养殖健康。近年来,杀鲑气单胞菌的感染病例越来越多,威胁多种海、淡水养殖鱼类。目前,杀鲑气单胞菌病原的鉴定和亚种区分主要依靠病原培养、生理生化检测和管家基因分析,如Long等(2020)运用生理生化检测、6个管家基因和16S rRNA系统发育分析等方法,鉴定了圆口铜鱼(Coreius guichenoti)中感染的杀鲑气单胞菌杀鲑亚种;刘宁等(2015)利用生化鉴定和16S rRNA序列分析等方法鉴定了细鳞鱼(Brachymystax lenok)中感染的杀鲑气单胞菌无色亚种;我国的国家标准(GB/T 15805.6-2008)也采用了生理生化特征为检测指标,作为鉴定杀鲑气单胞菌和区分亚种的标准。但生理生化实验需要经过微生物纯化和培养过程,用时长,且部分新发现的临床菌株生理生化反应并不完全符合亚种鉴定标准,给鉴定带来了一定的困难。16S rRNA测序是细菌鉴定和系统发育研究的最有力方法(李志刚等, 2001),对区分大多数气单胞菌的分类具有重要参考价值(Martinez-Murcia et al, 1992),但测序会增加病原检测和鉴定的周期,且无法区分杀鲑气单胞菌亚种。
在杀鲑气单胞菌的快速检测方面,PCR技术已经得到了广泛应用,如常规PCR (Beaz-Hidalgo et al, 2008)、多重PCR (Onuk et al, 2010)、实时荧光定量PCR (Bartkova et al, 2017; 刘帅等, 2017)等。其中,vapA基因编码A层蛋白(A-layer protein)是杀鲑气单胞菌的重要毒力蛋白,vapA基因也是杀鲑气单胞菌鉴定和分型的重要分子标记(Lago et al, 2012)。通过对vapA基因序列的进化发育分析,可以将杀鲑气单胞菌分为14个亚型以及缺乏vapA基因的溶果胶亚种(Gulla et al, 2019)。Gustafson等(1992)建立了基于vapA基因的杀鲑气单胞菌PCR检测方法,检测下限为5 fg。我国出入境检验检疫行业标准(SN/T 2095- 2010)也以vapA基因为靶点进行PCR检测来鉴定杀鲑气单胞菌。但目前大多数杀鲑气单胞菌的特异性PCR检测方法只能检测到种,无法确定亚种,亚种鉴定还需要依靠生理生化分析和基因序列进化分析。
不同的杀鲑气单胞菌亚种在毒力、宿主偏好性和疫苗交叉免疫等方面均有差异,因此,确定其亚种对于疾病的诊断、治疗和预防具有重要意义。目前,杀鲑气单胞菌亚种的快速检测方法非常少,只有2种针对杀鲑亚种的检测(Miyata et al, 1996; Fernández-Álvarez et al, 2016),但随着更多的菌株基因组测序,通过GenBank中最新的基因组序列信息分析发现,这2种方法在检测中均存在一定的不足,可能漏检某些地理区域的临床株,或发生非特异性扩增。本研究针对杀鲑气单胞菌杀鲑亚种phoB基因和杀日本鲑亚种LOC111476736基因设计了引物A. salmonicida F1/R1和A. masoucida F2/R2,可以特异性地检测杀鲑气单胞菌的杀鲑亚种和杀日本鲑亚种。经过反应体系优化,该方法检测灵敏度在DNA水平上分别达到17.6和27.2 fg,在细胞水平达到12.8和23.8 CFU/反应,与Høie等(1997)以质粒序列为靶基因建立的PCR检测方法的检测灵敏度相当,具有较高的检测灵敏度。同时,本方法以23株水产常见的其他病原菌和环境菌株进行了特异性检测,结果显示,只有杀鲑气单胞菌杀鲑亚种和杀日本鲑亚种检测到特异性条带,其他细菌中无特异性条带,说明该检测方法具有较高的特异性。本方法也可以从人工感染发病鱼的组织中检测到杀鲑气单胞菌的2个亚种,具有较好的产业应用性。综上所述,本研究建立的杀鲑气单胞菌杀鲑亚种和杀日本鲑亚种的特异性检测方法具有较高的灵敏度和特异性,且应用方便,可作为流行病学调查和疾病快速诊断的一个重要依据。
ADAMS A, THOMPSON K. Development of an enzyme-linked immunosorbent assay (ELISA) for the detection of Aeromonas salmonicida in fish tissue. Journal of Aquatic Animal Health, 1990, 2(4): 281-288 DOI:10.1577/1548-8667(1990)002<0281:DOAELI>2.3.CO;2 |
ALTINOK I, CAPKIN E, KAYIS S. Development of multiplex PCR assay for simultaneous detection of five bacterial fish pathogens. Veterinary Microbiology, 2008, 131(3/4): 332-338 |
BARTKOVA S, KOKOTOVIC B, SKALL H F, et al. Detection and quantification of Aeromonas salmonicida in fish tissue by real-time PCR. Journal of Fish Diseases, 2017, 40(2): 231-242 DOI:10.1111/jfd.12505 |
BEAZ-HIDALGO R, MAGI G E, BALBOA S, et al. Development of a PCR protocol for the detection of Aeromonas salmonicida in fish by amplification of the fstA (ferric siderophore receptor) gene. Veterinary Microbiology, 2008, 128(3/4): 386-394 |
BYERS H, GUDKOVS N, CRANE M. PCR-based assays for the fish pathogen Aeromonas salmonicida. I. Evaluation of three PCR primer sets for detection and identification. Diseases of Aquatic Organisms, 2002, 49(2): 129-138 |
DEL CERRO A, MARQUEZ I, GUIJARRO J A. Simultaneous detection of Aeromonas salmonicida, Flavobacterium psychrophilum, and Yersinia ruckeri, three major fish pathogens, by multiplex PCR. Applied and Environmental Microbiology, 2002, 68(10): 5177-5180 DOI:10.1128/AEM.68.10.5177-5180.2002 |
FERNÁNDEZ-ÁLVAREZ C, GONZÁLEZ S F, SANTOS Y. Development of a SYBR green I real-time PCR assay for specific identification of the fish pathogen Aeromonas salmonicida subspecies salmonicida. Applied Microbiology and Biotechnology, 2016, 100(24): 10585-10595 DOI:10.1007/s00253-016-7929-2 |
GULLA S, BAYLISS S, BJÖRNSDÓTTIR B, et al. Biogeography of the fish pathogen Aeromonas salmonicida inferred by vapA genotyping. FEMS Microbiology Letters, 2019, 366(7): 1-7 |
GUSTAFSON C E, THOMAS C J, TRUST T J. Detection of Aeromonas salmonicida from fish by using polymerase chain reaction amplification of the virulence surface array protein gene. Applied and Environmental Microbiology, 1992, 58(12): 3816-3825 DOI:10.1128/aem.58.12.3816-3825.1992 |
HINEY M P, SMITH P R. Validation of polymerase chain reaction-based techniques for proxy detection of bacterial fish pathogens: Framework, problems and possible solutions for environmental applications. Aquaculture, 1998, 162(1/2): 41-68 |
HØIE S, HEUM M, THORESEN O F. Evaluation of a polymerase chain reaction-based assay for the detection of Aeromonas salmonicida ss salmonicida in Atlantic salmon Salmo salar. Diseases of Aquatic Organisms, 1997, 30(1): 27-35 |
KEELING S, BROSNAHAN C, JOHNSTON C, et al. Development and validation of a real-time PCR assay for the detection of Aeromonas salmonicida. Journal of Fish Diseases, 2013, 36(5): 495-503 DOI:10.1111/jfd.12014 |
KULKARNI A, CAIPANG C M, BRINCHMANN M F, et al. Loop- mediated isothermal amplification-An assay for the detection of atypical furunculosis caused by Aeromonas salmonicida in Atlantic cod, Gadus morhua. Journal of Rapid Methods and Automation in Microbiology, 2009, 17(4): 476-489 DOI:10.1111/j.1745-4581.2009.00184.x |
LAGO E P, NIETO T P, FARTO R. Virulence factors of Aeromonas salmonicida subsp. salmonicida strains associated with infections in turbot Psetta maxima. Diseases of Aquatic Organisms, 2012, 99(2): 145-151 |
LEE C, CHO J C, LEE S H, et al. Distribution of Aeromonas spp. as identified by 16S rDNA restriction fragment length polymorphism analysis in a trout farm. Journal of Applied Microbiology, 2002, 93(6): 976-985 |
LI S W, WANG D, LIAN H M, et al. Isolation, identification and pathogenicity of Aeroonas salmonicida subsp. achromogenes from Atlantic salmon (Salmo salar). Acta Hydrobiologica Sinica, 2015, 39(1): 234-240 [李绍戊, 王荻, 连浩淼, 等. 大西洋鲑杀鲑气单胞菌无色亚种的分离鉴定和致病性研究. 水生生物学报, 2015, 39(1): 234-240] |
LI Z G, YANG G P, ZHU Y H. The relationship between polymorphism of bacterial 16S rDNA restriction fragment length and community structure of aquatic bacteria. Acta Hydrobiologica Sinica, 2001, 25(2): 111-115 [李志刚, 杨官品, 朱艳红. 水环境细菌16S rDNA限制性片段长度多型性及群落结构分析. 水生物学报, 2001, 25(2): 111-115] |
LIU N, SHI X, DU Y C, et al. Isolation and identification of Aeromonas salmonicida from diseased Brachymystax lenok. Freshwater Fisheries, 2015, 45(1): 88-92 [刘宁, 时晓, 杜迎春, 等. 患病细鳞鱼杀鲑气单胞菌的分离与鉴定. 淡水渔业, 2015, 45(1): 88-92 DOI:10.3969/j.issn.1000-6907.2015.01.017] |
LIU S, LU T Y, WANG D, et al. Establishment and application of SYBR Green Ⅰ real-time PCR assay for detection of Aeromonas salmonicida. Genomics and Applied Biology, 2017, 36(7): 2884-2891 [刘帅, 卢彤岩, 王荻, 等. 杀鲑气单胞菌SYBR GreenⅠ实时荧光定量PCR检测方法的建立与应用. 基因组学与应用生物学, 2017, 36(7): 2884-2891] |
LONG M, LI T T, JIANG Y, et al. Isolation and characterization of Aeromonas salmonicida subspecies salmonicida from largemouth bronze gudgeon (Coreius guichenoti) cage-cultured in the upper reaches of Yangtze River. Acta Hydrobiologica Sinica, 2020, 44(1): 153-161 |
MARTINEZ-MURCIA A J, BENLLOCH S, COLLINS M D. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: Lack of congruence with results of DNA-DNA hybridizations. International Journal of Systematic Bacteriology, 1992, 42(3): 412-421 DOI:10.1099/00207713-42-3-412 |
MIYATA A, INGLIS V, AOKI T. Rapid identification of Aeromonas salmonicida subspecies salmonicida by the polymerase chain reaction. Aquaculture, 1996, 141(1/2): 13-24 |
NAJIMI M, LEMOS M L, OSORIO C R. Identification of siderophore biosynthesis genes essential for growth of Aeromonas salmonicida under iron limitation conditions. Applied Environmental Microbiology, 2008, 74(8): 2341-2348 DOI:10.1128/AEM.02728-07 |
ONUK E E, CIFTCI A, FINDIK A, et al. Development and evaluation of a multiplex PCR assay for simultaneous detection of Flavobacterium psychrophilum, Yersinia ruckeri and Aeromonas salmonicida subsp. salmonicida in culture fisheries. Journal of Veterinary Science, 2010, 11(3): 235-241 |
PAVAN M E, ABBOTT S L, ZORZOPULOS J, et al. Aeromonas salmonicida subsp. pectinolytica subsp. nov., a new pectinase- positive subspecies isolated from a heavily polluted river. International Journal of Systematic and Evolutionary Microbiology, 2000, 50(3): 1119-1124 |
WANG X R, CHEN S Q, MO Z L, et al. Isolation and identification of Aeromonas salmonicida associated with furunculosis in cultured sablefish (Anoplopoma fimbria). Progress in Fishery Sciences, 2017, 38(5): 25-31 [王晓冉, 陈四清, 莫照兰, 等. 养殖裸盖鱼(Anoplopoma fimbria)疥疮病病原菌的分离与鉴定. 渔业科学进展, 2017, 38(5): 25-31] |
ZANG X J, FANG H, CHEN C Z, et al. Biological characterization and phylogenetic analysis of pathogenic new subspecies of Aeromonas salmonicida isolated from stone flounder (Kareius bicoloratus L). Journal of Fishery Sciences of China, 2006, 13(6): 917-923 [张晓君, 房海, 陈翠珍, 等. 杀鲑气单胞菌一新亚种的生物学特性及系统发育学分析. 中国水产科学, 2006, 13(6): 917-923] |
ZHOU D R, LUO Y Z, HANG X Y et al. Isolation and identification of pathogens Aeromonas salmonicida salmonicida subsp. from northern snakehead (Ophicephalus argus). Transactions of Oceanology and Limnology, 2015(3): 64-70 [周冬仁, 罗毅志, 杭小英, 等. 1株乌鳢源杀鲑气单胞菌杀鲑亚种的分离与鉴定. 海洋湖沼通报, 2015(3): 64-70] |



