 生长关键基因
生长关键基因
 (Leiocassis longirostris)生长相关基因,本研究运用Illumina高通量测序技术比较分析了快速生长组[平均体质量为(534.02±53.68) g]和缓慢生长组[平均体质量为(108.41±4.96) g]各9尾长吻
(Leiocassis longirostris)生长相关基因,本研究运用Illumina高通量测序技术比较分析了快速生长组[平均体质量为(534.02±53.68) g]和缓慢生长组[平均体质量为(108.41±4.96) g]各9尾长吻 的脑组织基因表达谱。测序共获得267 404 674个高质量测序片段(clean reads),通过2种不同生长速率长吻
的脑组织基因表达谱。测序共获得267 404 674个高质量测序片段(clean reads),通过2种不同生长速率长吻 脑组织转录组比较筛选出518个差异表达基因,其中,412个基因表达量上调,106个基因表达量下调。对12个差异表达基因进行实时荧光定量PCR验证的结果与转录组测序结果一致。GO功能分类显示,大量差异表达基因富集到生长(growth)、生长因子活性(growth factor activity)和激素介导的信号通路(hormone-mediated signaling pathway) GO条目中。KEGG富集分析显示,一些差异表达基因在MAPK信号通路(MAPK signaling pathway)、转化生长因子β信号通路(TGF-beta signaling pathway)、钙离子信号通路(calcium signaling pathway)和神经活性配体–受体相互作用(neuroactive ligand-receptor interaction)等途径中富集。根据GO功能注释和KEGG富集分析,筛选出gnrh、thr、egr1、fgf18、sst、gipr、cart和crf等基因是调控长吻
脑组织转录组比较筛选出518个差异表达基因,其中,412个基因表达量上调,106个基因表达量下调。对12个差异表达基因进行实时荧光定量PCR验证的结果与转录组测序结果一致。GO功能分类显示,大量差异表达基因富集到生长(growth)、生长因子活性(growth factor activity)和激素介导的信号通路(hormone-mediated signaling pathway) GO条目中。KEGG富集分析显示,一些差异表达基因在MAPK信号通路(MAPK signaling pathway)、转化生长因子β信号通路(TGF-beta signaling pathway)、钙离子信号通路(calcium signaling pathway)和神经活性配体–受体相互作用(neuroactive ligand-receptor interaction)等途径中富集。根据GO功能注释和KEGG富集分析,筛选出gnrh、thr、egr1、fgf18、sst、gipr、cart和crf等基因是调控长吻 生长发育的关键候选基因。本研究结果为后续深入研究长吻
生长发育的关键候选基因。本研究结果为后续深入研究长吻 生长调控机制提供了重要的参考资料。
生长调控机制提供了重要的参考资料。 鱼类生长 转录组测序 神经内分泌因子
鱼类生长 转录组测序 神经内分泌因子 生长是养殖水产动物最具经济价值的性状之一,生长性能高的养殖鱼类往往能在满足人类食物需求的同时带来直接的经济效益。与哺乳动物相似,鱼类生长包括了能量代谢和肌肉生长等多种过程,主要受环境、基因以及基因与环境相互作用的影响,是一个复杂的数量性状(Dai et al, 2015)。目前,研究者们从肌肉生成和肝脏代谢过程的角度出发,已挖掘出多种鱼类的生长相关基因和生长调控机制。如王兰梅等(2021)确定了mb、my12b和tnnil等6个基因为福瑞鲤2号(Cyprinus carpio)肌肉生长的关键基因;Li等(2022)初步证明禾花鲤(Cyprinus carpio)生长差异可能是由于蛋白质沉积引起肌纤维肥大所致,进而表明了泛素–蛋白酶体途径是影响禾花鲤生长的重要因素;Zhang等(2021)在草鱼(Ctenopharyngodon idella)肝脏组织中鉴定出的ghr、igf1和igf1r主要在PI3K-Akt和mTOR信号通路中参与生长调控。然而,下丘脑可直接或间接地对机体生理节律、摄食、繁殖和生长等生命活动进行调控,是代谢过程和内分泌活动的重要神经调节中枢(Piórkowska et al, 2020)。一些神经内分泌因子(GH、GnRH、NPY、THR、CCK和SST等)和神经调控轴也对鱼类的生长调节起着不可或缺的作用(Canosa et al, 2007、2020; Dai et al, 2015; Li et al, 2010; Christian et al, 2007; Peng et al, 1997)。因此,利用脑组织开展鱼类生长的研究有利于分析生长相关的神经内分泌调控网络及关键基因。
转录组测序技术有助于深入探究细胞中基因的转录和转录调控。近年来,转录组学技术已广泛应用于水产动物免疫应答(Xue et al, 2021)、生长发育(Liu et al, 2020)、生物进化(Schunter et al, 2014)和环境适应(Yao et al, 2021)等方面的研究,有效地进行了功能基因挖掘、特异性状主效基因搜索和基因表达调控等研究(Ye et al, 2018; 罗辉等, 2015)。转录组测序技术的运用在很大程度上满足了水产动物生长发育相关功能基因和调节机制研究的需要,已在多种鱼类中确定了与生长相关的候选基因及其表达模式(王兰梅等, 2021; Lu et al, 2020; Tian et al, 2020; Lin et al, 2019)。
长吻






实验用长吻

|
|
表 1 长吻 |

参照TRIzol试剂(Invitrogen, 美国)的操作说明提取18尾长吻
分别从2个组中随机选择3个个体RNA等量混合,通过带有Oligo (dT)的磁珠富集mRNA后,用Fragmentation Buffer将mRNA随机打断,再以片段化的mRNA为模板合成cDNA第一链和第二链。合成的双链cDNA经纯化后,先进行末端修复、加A尾并连接测序接头,再用AMPure XP beads筛选370~420 bp左右的cDNA片段进行PCR扩增并纯化扩增产物,最终构建长吻
测序获得的原始数据(raw reads)中包含少量带有接头或测序质量较低的reads,为保证转录组分析质量及有效性,需对原始数据进行过滤。具体包括:剔除由于测序仪器误差和人为因素导致的低质量reads (QPhred≤20的碱基数占整个read长度的50%以上);去除含N (无法确定碱基信息)比率超过10%的reads;识别并切除带有接头序列的reads。
1.5 基因功能注释和基因差异表达分析使用HISAT2(v2.0.5)软件将clean reads与长吻
选取12个差异倍数较大的差异表达基因进行qRT-PCR,验证测序结果的准确度,相关引物见表 2。参照PrimeScriptTM RT-PCR (TaKaRa)试剂盒说明书进行逆转录,获得对应cDNA,以β-actin为内参基因,在ABI QuantStudio 3 Real-Time PCR系统上进行,每个样品的技术重复均为3。反应体系为10 μL: 5 μL 2×TB Green Premix Ex Taq Ⅱ (TaKaRa),0.2 μL ROX Reference Dye Ⅱ (50×),3 μL灭菌水,1 μL cDNA模板和上下游引物各0.4 μL (10 μmol/L)。反应条件:预变性95 ℃ 30 s;95 ℃ 5 s,60 ℃ 34 s,40个循环;95 ℃ 15 s,60 ℃ 1 min,95 ℃ 15 s。
|
|
表 2 验证所用引物信息 Tab.2 The information of primers used for validation |
体质量测定数据均以平均值±标准差(Mean±SD)表示,并采用SPSS 26.0软件进行独立样本T检验,P < 0.05表示差异极显著。qRT-PCR验证数据以2–ΔΔCt法计算基因的相对表达量,并采用SPSS 26.0软件对结果进行统计分析。
2 结果 2.1 长吻
所选18尾长吻



经转录组测序获得的FG和SG文库的raw reads分别为134 682 652和137 487 704。质控后获得的clean reads分别为132 315 488和135 089 186。碱基质量及组成分析显示,各组GC含量区间为45.04%~ 45.64%,各样品Q30的碱基质量值比例均大于92% (表 3),表明转录组测序数据质量高,可以用于后续分析。长吻
|
|
表 3 RNA-Seq数据统计 Tab.3 Summary of RNA-Seq data |
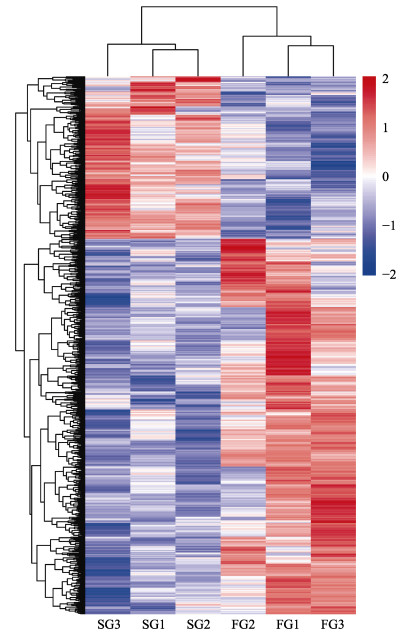
基于表达量指标FPKM,以P < 0.05、|log2(fold change)| > 1为阈值,对同一基因在FG组和SG组中的表达进行统计分析。与SG组相比,FG组中有412个基因表达量上调,106个基因表达量下调(图 1)。进一步对FG组和SG组间的518条差异基因进行层次聚类(hierarchical clustering)分析(图 2)。聚类结果显示,这些差异基因在2个比较组间的表达模式相差较大,而在组内不同样品间的表达模式比较相似。

|
图 1 长吻 |
|
图 2 差异表达基因聚类热图 Fig.2 Heat-map of differentially expressed genes 图中每行代表一个基因,每列代表一个样品;不同颜色区域分别代表不同的聚类分组信息,颜色由红到蓝表示差异表达基因的表达量由高到低。 In the heat-map, each row represents one gene and each column represents one sample; Different color areas represent different clustering information, the color from red to blue represents the expression intensity of differentially expressed genes from high to low. |
通过clusterProfiler (v3.8.1)软件对差异表达基因进行GO和KEGG富集分析,在GO功能分类体系中,518条差异表达基因共获得463个GO功能注释。其中,生物学过程类别(biological process, BP) 215个,细胞组分(cell composition, CC) 51个,分子功能类别(molecular function, MF) 197个。由前30个显著富集的GO terms可见,在生物学过程类型中,大量上调基因富集到免疫反应(immune response)、免疫系统过程(immune system process)和细胞死亡(cell death)等;细胞组分类别中,质膜部分(plasma membrane part)、细胞质膜(plasma membrane)和质膜蛋白复合物(plasma membrane protein complex)富集到的差异表达基因最多;涉及到分子功能的差异表达基因主要参与的生命过程有四吡咯结合(tetrapyrrole binding)、血红素结合(heme binding)以及辅因子结合(cofactor binding)等(图 3)。此外,有部分差异表达基因在生长(growth)、生长因子活性(growth factor activity)和激素介导的信号通路(hormone-mediated signaling pathway) GO terms中富集。

|
图 3 长吻 |
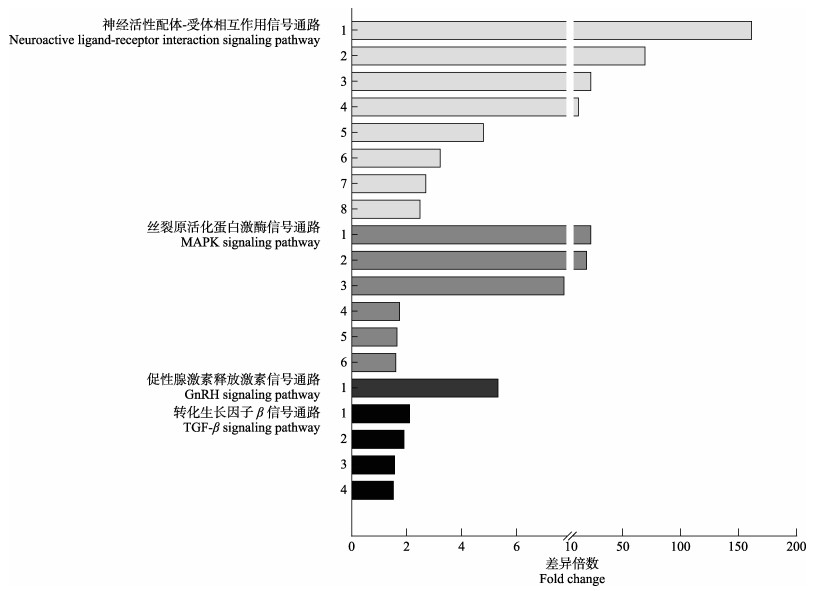
差异基因KEGG富集分析结果显示,长吻

|
图 4 长吻 |
参考脊椎动物生长信号通路调控模式和差异基因的功能,并根据差异表达基因的GO功能分类和KEGG富集分析,在相应的调控通路中初步筛选出19个与长吻
|
图 5 筛选到的长吻 |
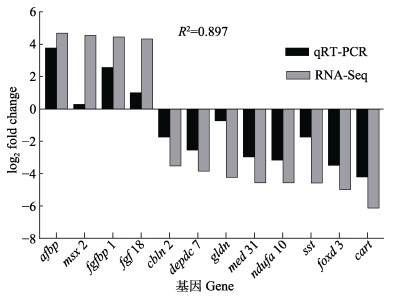
用于qRT-PCR验证的差异表达基因包括4个上调基因:脂肪细胞型脂肪酸结合蛋白、同源异型盒蛋白、成纤维细胞生长因子结合蛋白1和成纤维细胞生长因子18;8个下调基因:小脑肽2、含DEP结构域的蛋白7、神经胶质蛋白、转录中介复合物亚基31、NADH泛醌氧化还原酶亚基10、生长激素抑制素、叉头框转录因子D3以及可卡因–安非他明调节转录肽。qRT-PCR验证结果显示,这12个差异表达基因的表达趋势与转录组测序结果基本一致(图 6),说明RNA-Seq分析结果可信。
|
图 6 差异表达基因的转录组测序和qRT-PCR比较(β-actin为内参基因) Fig.6 Comparison of differentially expressed genes by RNA-Seq and qRT-PCR (β-actin was used as an internal gene) |
目前,运用转录组测序技术对水产动物生长发育进行的研究主要是针对肌肉生成和肝脏代谢过程。如Zhang等(2020)和Lu等(2020)分别构建了不同生长速率的青鱼和草鱼的肝脏以及肌肉转录组文库,筛选了几个与生长发育相关的关键基因和代谢途径;而对鱼类脑组织进行的转录组分析主要为了探究脑组织基因表达与生殖发育(Cardoso et al, 2018; Saaristo et al, 2017; Partridge et al, 2016)、生物学特性(Vu et al, 2021; Wei et al, 2021; Wang et al, 2020)以及环境适应能力(Bao et al, 2021; Feng et al, 2021; Zhang et al, 2020)的关系。鱼体的正常生理活动和生化过程是在神经系统的主导下实现的。脑作为中枢神经系统的重要组成部分,对神经内分泌轴上生长相关激素的合成和分泌具有不可替代的调控作用。除此之外,鱼类的生长涉及到复杂的调控网络,生长轴上的基因在调控鱼类的生长和代谢过程中扮演着关键的角色,而生长轴往往是处于脑的支配下参与鱼类生长过程。然而,目前鲜有通过鱼类脑组织挖掘生长相关基因的研究报道。现有研究主要有:Li等(2021)通过高通量测序,初步阐明了三倍体鲫鱼(Carassius auratus)生长快、抗病能力强的分子机制与基因表达水平升高密切相关;Robledo等(2017)分析了不同生长速率大菱鲆(Scophthalmus maximus)肌肉和脑组织基因表达谱,但在脑组织中只检测到几个差异表达基因,这些基因在先前的研究中被证实与鱼类感觉调控有关;Lin等(2021)对不同生长速率黑鲷(Acanthopagrus schlegelii)肝脏、肌肉和脑组织的混合样本进行了转录组测序,旨在挖掘生长相关候选基因和调控途径,但其结果尚不能说明黑鲷脑组织与其生长之间的关联。由此,仍需进一步明确鱼类脑组织与生长之间的关系和潜在的分子机制。因此,为探明长吻




鱼类的生长和发育受体内各种激素及其相互作用的调节,其中生长激素–胰岛素样生长因子轴(growth hormone-insulin like growth factor axis, GH-IGFs)是调控鱼类生长的内分泌核心(代向燕等, 2014),GH的合成和分泌是该过程的重要基础。在内分泌调节活动中,激素本身与相应受体匹配是其发挥作用的关键环节,促性腺激素释放激素(gonadotropin-releasing hormone, GnRH)和促甲状腺激素释放激素(thyrotropin-releasing hormone, TRH)主要是通过相应受体来介导并发挥生理功能。在本研究FG组和SG组脑组织的差异表达基因中,促性腺激素释放激素受体基因(gonadotropin- releasing hormone receptor, gnrhr)和促甲状腺激素释放激素受体基因(thyrotropin-releasing hormone receptor, trhr)高表达于FG组,提示可能有大量gnrh和trh与这些受体结合,并参与刺激长吻



除与生长激素调控相关的内分泌因子外,在差异表达基因中还筛选到了胰岛素样生长因子Ⅱ(insulin- like growth factor Ⅱ, igfⅡ)、成纤维细胞生长因子18 (fibroblast growth factor 18, fgf18)、成纤维细胞生长因子结合蛋白1 (fibroblast growth factor-binding protein 1, fgfbp1)和早期生长反应蛋白1 (early growth response protein, egr1)等生长相关基因以及NGFI-A结合蛋白2基因(NGFI-A-binding protein 2, nab2)。FGF家族nab2是egr1的特异性抑制剂,可通过抑制egr1的转录调控过程来有效减少egr1的表达(陈子翔, 2016)。egr1是转录因子锌指蛋白家族的重要成员之一,可正向调控igfⅡ和fgf的表达,以达到促进机体生长发育的作用(Liu et al, 1996)。本研究中,igfⅡ、fgf18和fgfbp1的表达趋势与egr1一致,均在FG组中上调表达,表明这几个基因可能是影响长吻


本研究还鉴定了胃抑制多肽受体基因(gastric inhibitory polypeptide receptor, gipr)、可卡因–苯丙胺调节转录肽基因(cocaine- and amphetamine-regulated transcript, cart)和促肾上腺皮质激素释放激素基因(corticotropin-releasing factor, crf)。相关研究指出,gipr基因作为肥胖、脂代谢紊乱及代谢综合征的易感基因(晋梦诗, 2014),可直接作用于脂肪组织,促进脂质沉积,gipr基因敲除或缺失的小鼠通过改变其能量消耗和脂肪代谢,以抵抗高脂饮食引起的肥胖(Boer et al, 2021; Miyawaki et al, 2002)。cart最早在大鼠弓状核中分离得到(Douglass et al, 1995),是一种可作用于下丘脑的厌食肽(Valassi et al, 2008),参与哺乳动物机体的进食行为和体重调节。相对于野生型小鼠,cart基因敲除小鼠在正常饮食条件下体重明显增加(Wierup et al, 2005)。该基因在金鱼中同样被证明是一种厌食因子(Hélène et al, 2000),在大西洋鲑(Salmo salar)中发挥抑制食欲的功能(Murashita et al, 2009)。Crf为CRF系统中的一员,可作用于下丘脑–垂体–肾上腺(hypothalamic-pituitary-adrenal, HPA)轴,刺激垂体释放促肾上腺皮质激素(adrenocorticotropic hormone, ATCH)进而调控动物的摄食行为(齐锦雯等, 2018)。在对金鱼(De Pedro et al, 1993)和虹鳟(Ortega et al, 2013)的研究中,发现注射CRF后实验鱼的摄食量减少,推测CRF可能通过中枢调控影响鱼类的摄食行为。Wang等(2014)发现,齐口裂腹鱼(Schizothorax prenanti)在长期禁食(7 d)条件下,其下丘脑crf基因的表达量明显下降,复食后则回升,表明crf可能作为厌食欲因子调控鱼类摄食。此外,Smith等(2004)对大鼠的研究表明crf可上调cart参与厌食欲作用。因此,推测crf可能同样可与cart互作共同调控鱼类的摄食行为。本研究中,gipr在FG组长吻


值得关注的是,我们发现参与生长调节的激素基因大多在神经活性配体–受体相互作用(neuroactive ligand-receptor interaction)和GnRH信号通路(GnRH signaling pathway)中富集,表明这些基因可能作为神经内分泌调节因子,对长吻




BAO M, SHANG F, LIU F, et al. Comparative transcriptomic analysis of the brain in Takifugu rubripes shows its tolerance to acute hypoxia. Fish Physiology and Biochemistry, 2021, 47(5): 1669-1685 DOI:10.1007/s10695-021-01008-6 |
BOER G A, KEENAN S N, MIOTTO P M, et al. GIP receptor deletion in mice confers resistance to high-fat diet-induced obesity via alterations in energy expenditure and adipose tissue lipid metabolism. AJP-Endocrinology and Metabolism, 2021, 320(4): E835-E845 DOI:10.1152/ajpendo.00646.2020 |
BRAZEAU P, VALE W, BURGUS R, et al. Hypothalamic polypeptide that inhibits the secretion of immunoreactive pituitary growth hormone. Science, 1973, 179(4068): 77-79 DOI:10.1126/science.179.4068.77 |
Bureau of Fisheries, Ministry of Agriculture and Rural Affairs, National Fisheries Technology Extension Center, China Society of Fisheries. China fishery statistical yearbook 2021. Beijing: China Agriculture Press, 2021 [农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会. 2021中国渔业统计年鉴. 北京: 中国农业出版社, 2021]
|
CANOSA L F, BERTUCCI J I. Nutrient regulation of somatic growth in teleost fish: The interaction between somatic growth, feeding and metabolism. Molecular and Cellular Endocrinology, 2020, 518: 111029 DOI:10.1016/j.mce.2020.111029 |
CANOSA L F, CHANG J P, PETER R E. Neuroendocrine control of growth hormone in fish. General and Comparative Endocrinology, 2007, 151(1): 1-26 DOI:10.1016/j.ygcen.2006.12.010 |
CARDOSO S D, GONÇALVES D, GOESMANN A, et al. Temporal variation in brain transcriptome is associated with the expression of female mimicry as a sequential male alternative reproductive tactic in fish. Molecular Ecology, 2018, 27(3): 789-803 DOI:10.1111/mec.14408 |
CHEN Z X. Investigation of the correlation between the expression of Egr-1, Nab2 and cav-1 and scar hyperplasia. Masterxs Thesis of Guangxi Medical University, 2016 [陈子翔. 探讨Egr-1、Nab2和cav-1的表达与瘢痕增生的相关性研究. 广西医科大学硕士研究生学位论文, 2016]
|
CHRISTIAN D S, DEAN R J. Candidate growth genes in finfish—Where should we be looking?. Aquaculture, 2007, 272(1/2/3/4): 22-38 |
DAI X Y, ZHANG W, ZHOU Z J, et al. Neuroendocrine regulation of somatic growth in fishes. Science China Life Sciences, 2015, 58(2): 137-147 DOI:10.1007/s11427-015-4805-8 |
DAI X Y, ZHANG W, ZHUO Z J, et al. Neuroendocrine control of fish growth. Scientia Sinica (Vitae), 2014, 44(12): 1213-1226 [代向燕, 张玮, 卓子见, 等. 鱼类生长的神经内分泌调控. 中国科学: 生命科学, 2014, 44(12): 1213-1226] |
DE PEDRO N, ALONSO-GÓMEZ A L, GANCEDO B, et al. Role of corticotropin-releasing factor (CRF) as a food intake regulator in goldfish. Physiology and Behavior, 1993, 53(3): 517-520 DOI:10.1016/0031-9384(93)90146-7 |
DONG X L, LEI W, ZHU X M, et al. Effects of dietary oxidized fish oil on growth performance and skin colour of Chinese longsnout catfish (Leiocassis longirostris Günther). Aquaculture Nutrition, 2011, 17(4): e861-e868 DOI:10.1111/j.1365-2095.2011.00854.x |
DOUGLASS J, MCKINZIE A, COUCEYRO P. PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. Journal of Neuroscience, 1995, 15(3): 2471-2481 DOI:10.1523/JNEUROSCI.15-03-02471.1995 |
FENG C, LI X, SHA H, et al. Comparative transcriptome analysis provides novel insights into the molecular mechanism of the silver carp (Hypophthalmichthys molitrix) brain in response to hypoxia stress. Comparative Biochemistry and Physiology Part D: Genomics and proteomics, 2021, 41: 100951 |
FONTAINE R, CIANI E, HAUG T M, et al. Gonadotrope plasticity at cellular, population and structural levels: A comparison between fishes and mammals. General and Comparative Endocrinology, 2020, 287: 113344 DOI:10.1016/j.ygcen.2019.113344 |
HAN D, XIE S Q, LEI W, et al. Effect of light intensity on growth, survival and skin color of juvenile Chinese longsnout catfish (Leiocassis longirostris Günther). Aquaculture, 2005, 248(1/2/3/4): 299-306 |
HAN D, XIE S Q, ZHU X M, et al. Physiological responses of Chinese longsnout catfish to water temperature. Chinese Journal of Oceanology and Limnology, 2011, 29(3): 633-639 DOI:10.1007/s00343-011-0165-9 |
HE W P, ZHOU J, LI Z, et al. Chromosome-level genome assembly of the Chinese longsnout catfish Leiocassis longirostris. Zoological Research, 2021, 42(4): 417-422 DOI:10.24272/j.issn.2095-8137.2020.327 |
HÉLÈNE V, PETER R E. Effects of cart peptides on food consumption, feeding and associated behaviors in the goldfish, Carassius auratus: Actions on neuropeptide Y- and orexin A-induced feeding. Brain Research, 2000, 887(1): 125-133 DOI:10.1016/S0006-8993(00)03001-8 |
JIN M S. Association study of GIP and gipr gene polymorphism with abdominal obesity and metabolic syndrome. Masterxs Thesis of Zhejiang University, 2014 [晋梦诗. GIP及gipr基因多态性与腹型肥胖及代谢综合征的相关性研究. 浙江大学硕士研究生学位论文, 2014]
|
LI S N, ZHOU Y, YANG C H, et al. Comparative analyses of hypothalamus transcriptomes reveal fertility-, growth-, and immune-related genes and signal pathways in different ploidy cyprinid fish. Genomics, 2021, 113(2): 595-605 DOI:10.1016/j.ygeno.2021.01.004 |
LI W S, LIN H R. The endocrine regulation network of growth hormone synthesis and secretion in fish: Emphasis on the signal integration in somatotropes. Science China Life Sciences, 2010, 53(4): 462-470 DOI:10.1007/s11427-010-0084-6 |
LI Z, DU X S, WEN L T, et al. Transcriptome analysis reveals the involvement of ubiquitin-proteasome pathway in the regulation of muscle growth of rice flower carp. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 2022, 41: 100948 DOI:10.1016/j.cbd.2021.100948 |
LI Z, JING T S, LI Y, et al. Effects of morphological traits on the body mass of Leiocassis longirostris. Progress in Fishery Sciences, 2021, 42(4): 98-105 [李哲, 敬庭森, 李雨, 等. 长吻  形态性状对体质量的影响. 渔业科学进展, 2021, 42(4): 98-105] 形态性状对体质量的影响. 渔业科学进展, 2021, 42(4): 98-105] |
LIN G, THEVASAGAYAM N M, WAN Z Y, et al. Transcriptome analysis identified genes for growth and omega-3/-6 ratio in saline tilapia. Frontiers in Genetics, 2019, 10: 244 DOI:10.3389/fgene.2019.00244 |
LIN Z J, ZHANG Z Y, SOLBERG M F, et al. Comparative transcriptome analysis of mixed tissues of black porgy (Acanthopagrus schlegelii) with differing growth rates. Aquaculture Research, 2021, 52(11): 5800-5813 DOI:10.1111/are.15455 |
LIU C, CALOGERO A, RAGONA G, et al. EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-beta1 is postulated to account for this suppressor activity. Critical Reviews in Oncogenesis, 1996, 7(1/2): 101-125 |
LIU X G, ZENG S, LIU S, et al. Identifying the related genes of muscle growth and exploring the functions by compensatory growth in mandarin fish (Siniperca chuatsi). Frontiers in Physiology, 2020, 11: 553563 DOI:10.3389/fphys.2020.553563 |
LU X, CHEN H M, QIAN X Q, et al. Transcriptome analysis of grass carp (Ctenopharyngodon idella) between fast- and slow-growing fish. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 2020, 35: 100688 DOI:10.1016/j.cbd.2020.100688 |
LUO H, YE H, XIAO S J, et al. Application of transcriptomics technology to aquatic animals research. Journal of Fisheries of China, 2015, 39(4): 598-607 [罗辉, 叶华, 肖世俊, 等. 转录组学技术在水产动物研究中的运用. 水产学报, 2015, 39(4): 598-607] |
MIYAWAKI K, YAMADA Y, BAN N, et al. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nature Medicine, 2002, 8(7): 738 DOI:10.1038/nm727 |
MURASHITA K, KUROKAWA T, EBBESSON L, et al. Characterization, tissue distribution, and regulation of agouti-related protein (AgRP), cocaine- and amphetamine-regulated transcript (CART) and neuropeptide Y (NPY) in Atlantic salmon (Salmo salar). General and Comparative Endocrinology, 2009, 162(2): 160-171 DOI:10.1016/j.ygcen.2009.03.015 |
ORTEGA V A, LOVEJOY D A, BERNIER N J. Appetite-suppressing effects and interactions of centrally administered corticotropin-releasing factor, urotensin I and serotonin in rainbow trout (Oncorhynchus mykiss). Frontiers in Neuroscience, 2013, 7: 196 |
PARTRIDGE C G, MACMANES M D, ROSEMARY K, et al. Brain transcriptional profiles of male alternative reproductive tactics and females in bluegill sunfish. PLoS One, 2016, 11(12): 1-21 |
PEI Z H, XIE S Q, LEI W, et al. Comparative study on the effect of dietary lipid level on growth and feed utilization for gibel carp (Carassius auratus gibelio) and Chinese longsnout catfish (Leiocassis longirostris Günther). Aquaculture Nutrition, 2015, 10(4): 209-216 |
PENG C, PETER R E. Neuroendocrine regulation of growth hormone secretion and growth in fish. Zoological Studies, 1997, 36(2): 79-89 |
PIÓRKOWSKA K, ŻUKOWSKI K, POŁTOWICZ K, et al. Identification of candidate genes and regulatory factors related to growth rate through hypothalamus transcriptome analyses in broiler chickens. BMC Genomics, 2020, 21(1): 1-12 DOI:10.1186/s12864-019-6419-1 |
QI J W, WU Y B, WANG S Y, et al. Corticotropin releasing factor: Suppression effects on fish feed intake. Chinese Journal of Animal Nutrition, 2018, 30(4): 1279-1285 [齐锦雯, 吴源冰, 王书瑶, 等. 促皮质激素释放激素对鱼类摄食的抑制作用. 动物营养学报, 2018, 30(4): 1279-1285 DOI:10.3969/j.issn.1006-267x.2018.04.010] |
ROBLEDO G, RUBIOLO J A, CABALEIRO S, et al. Differential gene expression and SNP association between fast- and slow-growing turbot (Scophthalmus maximus). Scientific Reports, 2017, 7(1): 12105 DOI:10.1038/s41598-017-12459-4 |
SAARISTO M, WONG B M, MINCARELLI L, et al. Characterisation of the transcriptome of male and female wild-type guppy brains with RNA-Seq and consequences of exposure to the pharmaceutical pollutant, 17 alpha-ethinyl estradiol. Aquatic Toxicology, 2017, 186: 28-39 DOI:10.1016/j.aquatox.2017.02.016 |
SCHUNTER C, VOLLMER S V, MACPHERSON E, et al. Transcriptome analyses and differential gene expression in a non-model fish species with alternative mating tactics. BMC Genomics, 2014, 15(1): 167 DOI:10.1186/1471-2164-15-167 |
SMITH S M, VAUGHAN J M, DONALDSON C J, et al. Cocaine- and amphetamine-regulated transcript activates the hypothalamic-pituitary-adrenal axis through a corticotropin-releasing factor receptor-dependent mechanism. Endocrinology, 2004, 145(11): 5202-5209 DOI:10.1210/en.2004-0708 |
SU J Z, MEI L Y, XI L W, et al. Responses of glycolysis, glycogen accumulation and glucose-induced lipogenesis in grass carp and Chinese longsnout catfish fed high-carbohydrate diet. Aquaculture, 2020, 533: 736146 |
TIAN C X, LIN X H, SAETAN W, et al. Transcriptome analysis of liver provides insight into metabolic and translation changes under hypoxia and reoxygenation stress in silver sillago (Sillago sihama). Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 2020, 36: 100715 DOI:10.1016/j.cbd.2020.100715 |
VALASSI E. Neuroendocrine control of food intake. Nutrition Metabolism and Cardiovascular Diseases, 2008, 18(2): 158-168 DOI:10.1016/j.numecd.2007.06.004 |
VU T D, IWASAKI Y, OSHIMA K, et al. Data of RNA-seq transcriptomes in the brain associated with aggression in males of the fish Betta splendens. Data in Brief, 2021, 38: 107448 DOI:10.1016/j.dib.2021.107448 |
WANG L M, ZHU W B, FU J J, et al. De novo transcriptome analysis and comparison of the FFRC No.2 strain common carp Cyprinus carpio associated with its muscle growth. Journal of Fisheries of China, 2021, 45(1): 79-87 [王兰梅, 朱文彬, 傅建军, 等. 福瑞鲤2号不同生长速率个体肌肉组织转录组分析. 水产学报, 2021, 45(1): 79-87] |
WANG M, XU G, TANG Y, et al. Transcriptome analysis of the brain provides insights into the regulatory mechanism for Coilia nasusmigration. BMC Genomics, 2020, 21(1): 410 DOI:10.1186/s12864-020-06816-3 |
WANG T, ZHOU C W, YUAN D Y, et al. Schizothorax prenanti corticotropin-releasing hormone (CRH): Molecular cloning, tissue expression, and the function of feeding regulation. Fish Physiology and Biochemistry, 2014, 40(5): 1407-1415 DOI:10.1007/s10695-014-9935-6 |
WEI Y L, WEN B, GAO J Z, et al. Brain transcriptome analysis reveals genes involved in parental care behavior in discus fish (Symphysodon haraldi). General and Comparative Endocrinology, 2021, 309: 113793 DOI:10.1016/j.ygcen.2021.113793 |
WIERUP N, RICHARDS W G, BANNON A W, et al. CART knock out mice have impaired insulin secretion and glucose intolerance, altered beta cell morphology and increased body weight. Regulatory Peptides, 2005, 129(1/2/3): 203-211 |
XIAO M S, YANG G. Isolation and characterization of 17 microsatellite loci for the Chinese longsnout catfish (Leiocassis longirostris). Molecular Ecology Resources, 2009, 9(3): 1039-1041 DOI:10.1111/j.1755-0998.2009.02554.x |
XUE T, LIU Y P, CAO M, et al. Transcriptome analysis reveals deep insights into the early immune response of turbot (Scophthalmus maximus) induced by inactivated Aeromonas salmonicida vaccine. Fish and Shellfish Immunology, 2021, 119: 163-172 DOI:10.1016/j.fsi.2021.09.027 |
YAO Z L, CHEN H J, ZHAO Y, et al. A time course transcriptome analysis of brains from sex-undifferentiated Nile tilapia discloses genes associated with high-temperature-induced masculinization. Aquaculture, 2021, 530: 735762 DOI:10.1016/j.aquaculture.2020.735762 |
YE H, LIN Q S, LUO H. Applications of transcriptomics and proteomics in understanding fish immunity. Fish and Shellfish Immunology, 2018, 77: 319-327 DOI:10.1016/j.fsi.2018.03.046 |
ZHANG J H, SHEN Y B, XU X Y, et al. Transcriptome analysis of the liver and muscle tissues of black carp (Mylopharyngodon piceus) of different growth rates. Marine Biotechnology, 2020, 22(5): 706-716 DOI:10.1007/s10126-020-09994-z |
ZHANG L, LI X H, YU Y L, et al. Comparative analyses of liver transcriptomes reveal the effect of exercise on growth-, glucose metabolism-, and oxygen transport-related genes and signaling pathways in grass carp (Ctenopharyngodon idella). Comparative Biochemistry and Physiology Part A: Molecular and Integrative Physiology, 2021, 262: 111081 DOI:10.1016/j.cbpa.2021.111081 |
ZHANG S L, SUN X J, ZHANG X, et al. The dressing rate and nutrient components in muscle of Leiocassis longirostris. Journal of Dalian Ocean University, 2013, 28(1): 83-88 [张升利, 孙向军, 张欣, 等. 长吻  含肉率及肌肉营养成分分析. 大连海洋大学学报, 2013, 28(1): 83-88] 含肉率及肌肉营养成分分析. 大连海洋大学学报, 2013, 28(1): 83-88] |
ZHANG Y, ZHANG P J, YU P, et al. Transcriptome analysis reveals the mechanism of fluorine exposure on memory loss of common carp. Environmental Pollution, 2020, 265(Pt A): 114927 |
ZHAO H Y, HAN D, XIE S Q, et al. Effect of water temperature on the growth performance and digestive enzyme activities of Chinese longsnout catfish (Leiocassis longirostris Günther): Temperature on growth and digestive enzyme. Aquaculture Research, 2009, 40(16): 1864-1872 DOI:10.1111/j.1365-2109.2009.02292.x |
ZHAO W, ZHOU J, LI Z, et al. Characterization of 55 SNP markers in Chinese longsnout catfish Leriocassis longirostris. Conservation Genetics Resources, 2020, 12(3): 427-432 DOI:10.1007/s12686-020-01137-9 |
ZHU X M, XIE S Q, LEI W, et al. Compensatory growth in the Chinese longsnout catfish, Leiocassis longirostris following feed deprivation: Temporal patterns in growth, nutrient deposition, feed intake and body composition. Aquaculture, 2005, 248(1/2/3/4): 307-314 |



