2. 中国科学院大学 北京 100049
2. University of Chinese Academy of Sciences, Beijing 100049, China
裙带菜(Undaria pinnatifida)是一种重要的经济海藻,生活史由微型的配子体和大型的孢子体交替组成,其栽培主要集中在中国、韩国和日本。据2020年中国渔业统计年鉴报道,我国裙带菜年产量超过20万t,在经济褐藻中仅次于海带(农业农村部渔业渔政管理局等, 2020)。与海带和紫菜等其他经济海藻相似,由于养殖面积和密度的增加以及渔业环境的恶化,裙带菜病害时有发生,造成养殖产量下降和经济损失。引发藻类病害的原因有附生微生物、附生藻类、敌害生物、环境因素和管理不当等(杨慧超等, 2019),其中由细菌等微生物引发的病害最为常见。
Bell等(1972)提出“藻际微环境”的概念,即藻类在生长的过程中向周围环境释放出有机物质,在藻体周围形成一种特殊的藻际环境,吸引微生物附着在藻体表面。藻类和附生微生物之间关系密切,藻类为附生细菌提供生长所需的有机物,附生细菌通过代谢产物为藻类提供营养物质和生长因子(Singh et al, 2014),调控海藻的发育和生长,某些细菌还可以帮助海藻抵抗海洋环境中致病细菌的侵袭(姜健等, 2005)。当藻际微环境维持动态平衡时,二者相互利用,共同生长发育;当二者关系失衡,例如,藻体受到损伤或环境因子发生改变时,菌群结构就可能发生变化,造成藻类的病害(Egan et al, 2014; 梁珊珊, 2019)。研究表明,附生微生物与海带、紫菜等经济海藻的烂苗脱苗病(陈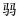
附生微生物的群落组成与多种因素有关,裙带菜配子体世代(n)和孢子体世代(2n)在形态生理上的不同决定了其附生微生物的群落可能存在差异。随着高通量测序技术的发展,以微生物基因组DNA的标记序列作为分类依据来分析微生物种群结构和多样性成为目前的常用方法。核糖体RNA操纵子在细胞中相对稳定,同时含有保守序列和高可变序列,是研究微生物系统分类的重要依据(车玉伶等, 2005)。16S rRNA基因是编码原核生物核糖体小亚基RNA的DNA序列,常用于研究细菌多样性;18S rRNA基因是编码真核生物核糖体小亚基RNA的DNA序列,常用于真核生物类群的研究。
本研究利用Illumina HiSeq 2500高通量测序平台,通过16S rRNA和18S rRNA基因高通量测序对裙带菜孢子体和配子体的附生微生物类群进行鉴别,分析比较不同世代裙带菜附生细菌及其他生物群落的组成和结构,为研究裙带菜与附生微生物间的相互作用、种质资源的有效保存以及防治藻类病害提供参考。
1 材料与方法 1.1 实验材料实验所用的裙带菜配子体和孢子体均来源于大连栽培群体,各3个生物学重复。其中,雌性、雄性和雌雄同体配子体各1个,编号分别为HB1-21-2F、HB2-15-1M和7-3-3FM (标记为G1、G2和G3)。它们为单个孢子衍生的克隆培养系,分别来自于“海宝1号”、“海宝2号”品种及“7号”品系,其建立时间均为2017年6月。保存条件为温度16 ℃、光照 < 5 μmol photons/(m2∙s),所用培养基为用煮沸的自然海水配制的PES (Provasoli enriched seawater)(Provasoli, 1968)。取样时,配子体为黄褐色,处于营养生长状态,培育密度约为1 g/100 mL(鲜重)。孢子体为“海宝1号”的3个个体(标记为S1、S2和S3)。孢子体为在水缸中培养的“海宝1号”的子代,培养基为添加70 mg/L NaNO3和10 mg/L NaH2PO4的自然海水,取样时海水的温度为16~18 ℃,光强的日变化在0~120 μmol photons/(m2∙s)。孢子体大小为15~20 cm,色泽黄褐色,取叶片中部用于后续的DNA提取。
1.2 建库测序用磁珠法土壤和粪便基因组DNA提取试剂盒(天根生化科技有限公司, DP712)提取样品总DNA,进行16S/18S rRNA基因的测序和微生物多样性基本分析。其中,16S rRNA基因的测序区间为V3~V4区,18S rRNA基因的测序区间为V4区。细菌16S rRNA基因使用分析海藻附生菌群常用的引物338F:5′-ACTCCTACGGGAGGCAGCA-3′和806R:5′-GGACTACHVGGGTWTCTAAT-3′(Hong et al, 2015; 孙丕海等, 2017; 唐隆晨等, 2020),18S rRNA基因v4的引物为TAReuk454FWD1:5′-CCAGCA(G/C)C(C/T)GCGGTAATTCC-3′和TAReukREV3:5′-ACTTTCGTTCTTGAT(C/T)(A/G)A-3′ (Stoeck et al, 2010)。在引物末端加上测序接头,进行PCR扩增,对扩增产物进行纯化、定量和均一化形成测序文库。质检合格的文库用Illumina HiSeq 2500进行双末端测序。
1.3 数据分析使用Trimmomatic v0.33软件(Bolger et al, 2014)对测序得到的原始序列(raw reads)进行过滤;使用cutadapt 1.9.1 (Martin, 2011)去除引物序列,使用FLASH v1.2.7 (Magoč et al, 2011),通过重叠区域对序列进行拼接,得到clean reads;使用UCHIME v4.2 (Edgar et al, 2011)去除嵌合体序列,得到有效序列。
使用Usearch v10.0软件(Edgar, 2013)对获得的有效序列在97.0%的相似度水平下进行聚类,获得可操作分类单元(operational taxonomic unit, OTU),每个OTU对应一种代表序列。以SILVA132为参考数据库,使用朴素贝叶斯分类器对特征序列进行分类学分析。使用QIIME2 2020.6 (Bolyen et al, 2019)对样品进行Alpha和Beta多样性分析。
2 结果与分析 2.1 16S rRNA基因序列分析结果 2.1.1 有效序列及OTU所取的6个裙带菜样品经测序后共获得460 754对原始序列,序列已上传至美国国家生物技术信息中心数据库(https://www.ncbi.nlm.nih.gov/),序列号为PRJNA823903。经双端质控、拼接后共产生455 735条clean reads,过滤嵌合体后得到有效序列共446 932条,在97.0%的相似度水平下聚类获得OTU,配子体样品的平均OTU数目(259)多于孢子体(110)(表 1)。
|
|
表 1 16S rRNA基因测序结果分析 Tab.1 Analysis of sequencing results of 16S rRNA gene |
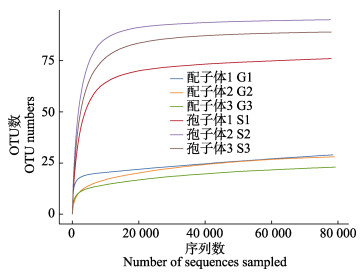
从测序样本中随机抽取一定数量的序列,统计抽取序列包含的OTU数量构建稀释性曲线(图 1)。随着有效序列数目的增加,各组样品的稀释曲线逐渐趋于平缓,表明获得的序列充分,可以较好地反映样品中细菌的多样性。
|
图 1 16S rRNA基因测序的稀释性曲线 Fig.1 Rarefaction curves of 16S rRNA gene sequencing |
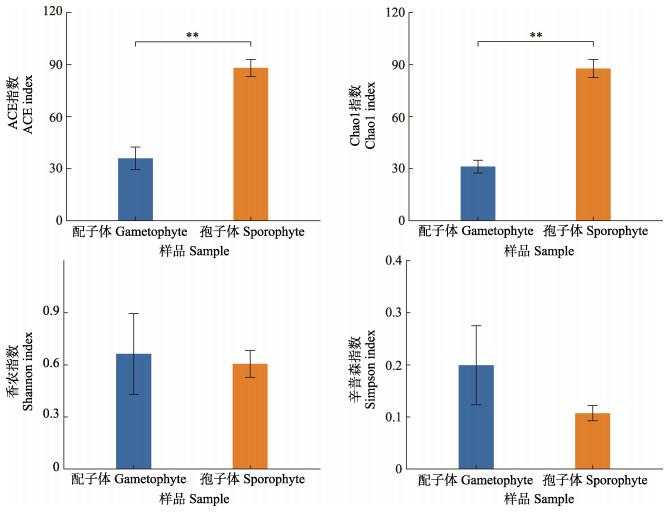
通过测序样品的ACE指数、Chao1指数、Shannon指数、Simpson指数和覆盖率来描述Alpha多样性(表 2)。各组测序结果的覆盖率均达到99%以上,说明测序结果能较好地反映样品中细菌群落的真实情况;ACE和Chao1指数能反映物种丰富度,裙带菜配子体样品的ACE和Chao1指数均显著高于孢子体样品(P < 0.01),说明配子体样品中细菌群落丰度大于孢子体;Shannon和Simpson指数用于衡量物种多样性(Grice et al, 2009),受群落中物种丰富度和物种均匀度影响,在物种丰度相同的情况下,群落中各物种均匀度越大,群落的多样性越大,配子体的Shannon和Simpson指数显著大于孢子体(P < 0.01),说明配子体样品的细菌多样性也大于孢子体(图 2)。
|
|
表 2 细菌群落的Alpha多样性指数 Tab.2 Alpha-diversity index of bacterial communities |

|
图 2 配子体与孢子体样品细菌群落的Alpha多样性指数比较 Fig.2 Comparisons of alpha-diversity index of bacterial communities between gametophytes and sporophytes **代表组间差异显著(P < 0.01),下同。 ** indicates significant difference between two groups (P < 0.01), the same as below. |
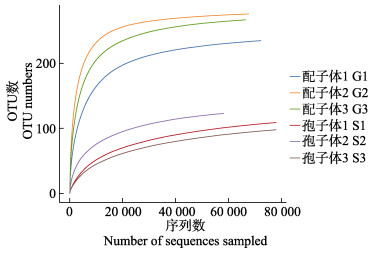
通过主坐标分析(principal coordinates analysis, PCoA)对裙带菜配子体和孢子体细菌群落结构的相似度进行比较分析。基于Weighted-Unifrac算法的Beta多样性分析结果见图 3,如图 3所示,孢子体组的3个样品明显聚集,与配子体组差异明显,PC1差异占总变异的92.00%,PC2差异占总变异的5.68%。
|
图 3 基于Weighted-Unifrac算法的配子体及孢子体细菌群落PCoA分析 Fig.3 PCoA result of bacterial communities in gametophytes and sporophytes based on Weighted-Unifrac algorithm |
裙带菜配子体和孢子体样品的共有OTU数目为145,配子体的独有OTU数目为149,孢子体的独有OTU数目为7。
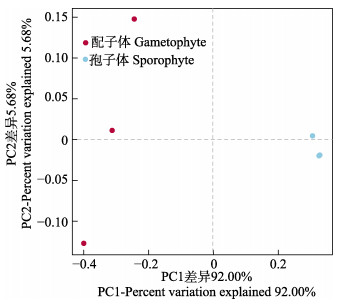
在门水平上,裙带菜附着细菌的主要类别是蓝细菌门(Cyanobacteria)、变形菌门(Proteobacteria)、拟杆菌门(Bacteroidetes)、厚壁菌门(Firmicutes)、放线菌门(Actinobacteria)、浮霉菌门(Planctomycetes)、疣微菌门(Verrucomicrobia)和绿弯菌门(Chloroflexi)(图 4A)。在配子体附着细菌中,变形菌门(66.67%)为第一优势菌门,其次为拟杆菌门(13.48%)、蓝细菌门(11.13%);而在孢子体中,蓝细菌门(95.67%)为第一优势菌门,其次为放线菌门(1.65%)和厚壁菌门(1.48%)。
|
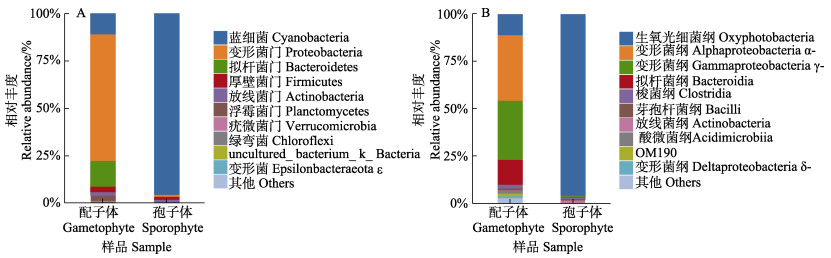
图 4 门(A)和纲(B)分类水平细菌类群分布 Fig.4 Distribution of bacterial communities on phylum (A)and class(B)levels |
在纲分类水平上,裙带菜附生细菌群落主要由生氧光细菌纲(Oxyphotobacteria)、α-变形菌纲(Alphaproteobacteria)、γ-变形菌纲、拟杆菌纲(Bacteroidia)、梭菌纲(Clostridia)、放线菌纲(Actinobacteria)和芽孢杆菌纲(Bacilli)组成(图 4B)。在配子体表面细菌群落中,α-变形菌纲(34.58%)为第一优势菌纲,其次为γ-变形菌纲(31.01%)、拟杆菌纲(13.16%)、生氧光细菌纲(11.13%);而在所取的孢子体样品中,生氧光细菌纲(95.67%)为第一优势菌纲,其次为放线菌纲(1.64%)、芽孢杆菌纲(0.92%)、γ-变形菌纲(0.90%)。
生氧光细菌纲、α-变形菌纲、拟杆菌纲、γ-变形菌纲、Negativicutes、OM190、酸微菌纲(Acidimicrobiia)、丹毒丝菌纲(Erysipelotrichia)、浮霉菌纲(Planctomycetacia)、疣微菌纲(Verrucomicrobiae) 10个纲的丰度在裙带菜配子体和孢子体样品之间存在显著差异(P < 0.05),其中,OM190、酸微菌纲和浮霉菌纲是配子体独有。
配子体样品中检出的属水平未培养细菌占57.37%,已知的细菌类群有赖文氏菌属(Lewinella)(10.06%)、亮发菌属(Leucothrix)(5.99%)、亚硫酸杆菌属(Sulfitobacter)(4.06%)、双歧杆菌属(Bifidobacterium)(0.02%);孢子体样品中未培养细菌占95.68%,双歧杆菌属占1.41%。
2.2 18S rRNA基因序列分析结果 2.2.1 有效序列及OTU6个样品经18S rRNA基因高通量测序后共获得480 390对raw reads,经双端质控、拼接后共产生475 128条clean reads,过滤嵌合体后得到的有效序列共473 770条。在97.0%相似度水平下聚类获得OTU,配子体样品的平均OTU数(27)少于孢子体(87)(表 3)。各组样品的稀释性曲线趋于平缓(图 5),说明获得的序列充分。
|
|
表 3 18S rRNA基因测序结果分析 Tab.3 Analysis of sequencing results of 18S rRNA gene |
|
图 5 18S rRNA基因测序的稀释性曲线 Fig.5 Rarefaction curves of 18S rRNA gene sequencing |
Alpha多样性指数如表 4所示,各组测序结果的覆盖率均达到99.99%以上,裙带菜孢子体样品的ACE和Chao1指数均显著高于配子体样品(P < 0.01),说明孢子体样品中群落丰度大于配子体;配子体、孢子体样品的Shannon和Simpson指数差异不显著(P > 0.05)(图 6)。
|
|
表 4 真核生物群落的Alpha多样性指数 Tab.4 Alpha-diversity index of eukaryotic communities |
|
图 6 配子体与孢子体样品真核生物群落的Alpha多样性指数比较 Fig.6 Comparisons of alpha-diversity index of eukaryotic communities between gametophytes and sporophytes |
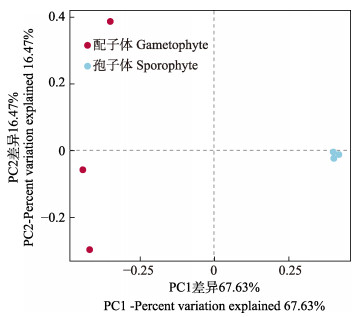
基于Binary-Jaccard算法的PCoA分析结果显示,孢子体组的3个样品明显聚集,PC1差异占总变异的67.63%,PC2差异占总变异的16.47% (图 7)。
|
图 7 基于Binary-Jaccard算法的配子体及孢子体真核生物类群PCoA分析 Fig.7 PCoA result of eukaryotic communities in gametophytes and sporophytes based on Binary-Jaccard algorithm |
裙带菜配子体和孢子体样品的共有OTU数目为22,配子体独有的OTU数目为23,孢子体独有OTU数目为87。
门分类水平测序结果显示(图 8A),配子体的97.22%、孢子体94.95%是未分类的真核微生物,此外检测出的占比较多的10个门是链形植物(Streptophyta)、纤毛虫门(Intramacronucleata)、担子菌门(Basidiomycota)、顶复亚门(Apicomplexa)、节肢动物门(Arthropoda)、硅藻门(Bacillariophyta)、脊索动物门(Chordata)、腹毛动物门(Gastrotricha)、子囊菌门(Ascomycota)和毛霉菌门(Mucoromycota),其中,担子菌门、子囊菌门和毛霉菌门属于真菌。配子体中链形植物、节肢动物门、纤毛虫门和硅藻门具有相对优势,占比分别为0.82%、0.62%、0.51%和0.50%;孢子体中链形植物、担子菌门、顶复亚门和纤毛虫门处于优势地位,所占OTUs比例分别为0.97%、0.96%、0.91%和0.78%。

|
图 8 门(A)和纲(B)水平真核生物类群分布 Fig.8 Distribution of eukaryotic communities on phylum(A)and class(B)levels |
在纲分类水平上(图 8B),在配子体样品中,除裙带菜所属的褐藻纲(Phaeophyceae)(88.77%)之外,节肢动物门的桡足亚纲(Copepoda)、硅藻门的间藻纲(Mediophyceae)、脊索动物中的哺乳纲(Mammalia)、纤毛虫门中的前口纲(Prostomatea)、百合纲(Liliopsida)处于优势地位,占比分别为0.62%、0.50%、0.25%、0.23%和0.23%。孢子体样品中,除褐藻纲(94.49%)之外,原生动物中的孢子虫纲(Conoidasida)、担子菌门中的伞菌纲(Agaricomycetes)、纤毛虫门中的旋唇纲(Spirotrichea)以及脊索动物门中的哺乳纲占优,分别占OTUs的0.91%、0.83%、0.51%和0.24%。
壶菌纲(Chytridiomycetes)、篮口纲(Nassophorea)、Colpodea、银耳纲(Tremellomycetes)、粪壳菌纲(Sordariomycetes)、类锥体纲(Conoidasida)、伞菌纲、蛛形纲(Arachnida)、色矛纲(Chromadorea) 9个纲仅存在于裙带菜孢子体样品中,旋唇纲的丰度在配子体和孢子体之间存在明显差异(P < 0.05)。
3 讨论本研究发现,裙带菜配子体和孢子体附生细菌群落结构差异明显,配子体样品中的细菌群落丰度和多样性均大于孢子体。附生细菌主要分布在蓝细菌门、变形菌门、拟杆菌门,此外,还有相对丰度较低的厚壁菌门、放线菌门、浮霉菌门、疣微菌门和绿弯菌门,这些菌门在海藻表面细菌群落中都有发现,且变形菌门通常占主导地位(孙丕海等, 2017; 唐隆晨等, 2020; 盖珊珊等, 2016; 武洪庆, 2012)。
已有研究证明,变形菌门中的某些附生细菌可以促进海藻的生长和发育,例如,α-变形菌纲中的生丝单胞菌(Hyphomonas sp.)可以降低条斑紫菜(Porphyra yezoensis)原生质体再生体的形态畸形率(Fukui et al, 2014);γ-变形菌纲中的一种嗜盐寡营养细菌(Halomonas sp.)能够在缺铁条件下加快杜氏藻(Dualiella bardawil)的生长速率(Keshtacher-Liebso et al, 1995);同属γ-变形菌纲的假单胞菌(Pseudomonas sp.)作为铜绿微囊藻(Microcystis aeruginosa)的附生细菌,能够将环境中不易吸收的磷形态转化为磷酸盐等物质供宿主利用,促进微囊藻生长(邹迪等, 2005);此外,拟杆菌门中的一些细菌类群对绿藻的生长发育和形态发生有影响(Marshall et al, 2006; Matsuo et al, 2003)。某些条件下,附生细菌还有可能导致藻类的病害,例如,假交替单胞菌(Pseudoalteromonas bacteriolytica sp.)是海带红斑病的病原菌(Sawabe et al, 1998);海带幼苗病烂的主要病原菌褐藻酸降解菌属于交替单胞菌属(Alteromonas Baumann)(王丽丽等, 2003);河豚毒素假交替单胞菌(Pseudoalteromonas tetraodonis)是条斑紫菜丝状体黄斑病的病原菌(王洪斌等, 2011)。由此可见,藻类附生细菌既能调控宿主的生长发育,特定条件下又可能导致病害。但大多数细菌在藻类生存中发挥的功能还没有详细的解释,藻类病害的发生又是一个复杂的过程,可能导致附生菌群结构发生变化的因素多种多样,因此,还需要针对附生细菌对宿主藻类发挥的作用及其机制进行深入研究。
与配子体不同,本研究孢子体样品中蓝细菌占绝对优势(95.67%),这一门类在厚成期海带的附生细菌中丰度也最高(唐隆晨等, 2020)。已有研究表明,具有固氮作用的蓝细菌参与宿主海带的氮同化过程,可能产生抵抗有害微生物的活性物质(Li et al, 2020),这可能是孢子体附生细菌种类较少的原因之一。另外,配子体与孢子体在形态、生长发育、代谢产物、存在的季节等诸多方面均存在明显差异,不同的生理特性以及不同的环境下的长期适应性进化也可能是其附生微生物存在较大差异的原因。相较于孢子体,配子体附生细菌种类更多,在实际的种质保存过程中,配子体为主要的保存对象,为了安全有效地进行种质保存,需要定期对配子体的生长状态进行显微观察,防止附生细菌大量繁殖对配子体造成不利影响。同时,本研究结果也为实现配子体的无菌克隆培养提供了基础的生物学依据。
OM190、酸微菌纲、浮霉菌纲3个纲仅存在于配子体,其中酸微菌纲在条斑紫菜贝壳丝状体(盖珊珊等, 2016)和海带表面(孙丕海等, 2017)都有发现。配子体样品中的亮发菌属在马泽藻(Mazzaella sp.)、海萝(Gloiopeltis furcate)、裙带菜附生菌群中都有发现,并且在马泽藻和裙带菜表面占优(武洪庆, 2012);亚硫酸杆菌属在海带表面曾被发现(孙丕海等, 2017)。这些细菌类群对海藻的意义尚未明确,有待进一步研究。
与附生细菌不同,孢子体的真核微生物群落丰度大于配子体。这可能与不同的培养环境有关,配子体所用培养基用经煮沸的海水配制,在煮沸的过程中一些真核生物可能被杀死,导致丰度降低。同时,蓝细菌是海洋环境中微食物网的重要部分,纤毛虫和异养鞭毛虫是其主要摄食者(孙晟等, 2005),孢子体中蓝细菌的大量存在可能吸引了捕食者和其他异养微生物,造成真核微生物种类的增加。
裙带菜经18S rRNA基因测序检出的微生物中有3种真菌,分别为担子菌门、子囊菌门和毛霉菌门。担子菌门和子囊菌门在坛紫菜藻际微生物中都有发现(沈梅丽等, 2013)。除褐藻纲之外,配子体中节肢动物门的桡足亚纲、硅藻门的间藻纲、脊索动物中的哺乳纲、纤毛虫门中的前口纲、百合纲处于优势地位;孢子体中,原生动物中的孢子虫纲、担子菌门中的伞菌纲、纤毛虫门中的旋唇纲以及脊索动物门中的哺乳纲占优。样品中检出的硅藻作为附生藻类,若大量繁殖会使紫菜生长受阻,造成经济损失(马家海, 1996b);某些纤毛虫以藻类为食,会造成藻类养殖产量的降低。徐润林等(2000)研究发现,前口纲在硅藻培养池中的有害纤毛虫中占优势,这一门类也存在于本研究中裙带菜配子体样品中。
本研究中,裙带菜配子体世代和孢子体世代的微生物群落组成和结构差异明显,配子体的附生菌群丰度和多样性均大于孢子体,真核微生物的丰度则小于孢子体。附生微生物的群落组成受多种生物和非生物因素影响。研究证实,不同生长阶段微藻产生的胞外聚合物组成不同(张圣洁等, 2020)。推测裙带菜不同世代向藻际微环境中释放的有机物成分和比例有所不同,导致了附生微生物群落的差异。此外,配子体和孢子体在形态和生理上的差异以及不同的培养环境,比如培养基成分和光照强度也会造成附生微生物群落差异。从PCoA分析可见,与孢子体样品的明显聚集不同,3个配子体样本间的微生物结构存在较大差异,其可能的原因有两个:第一,它们来自于不同的品种/系,在分离游孢子构建配子体克隆系之初,不同品种/系的微生物群落可能存在差异;第二,3个配子体性别的不同也可能造成其附生微生物存在差异。
本研究分析比较了裙带菜不同世代附生细菌及其他生物群落的组成和结构,为研究裙带菜与附生微生物间的相互作用、种质资源的有效保存以及防治藻类病害提供了基础数据支持。在后续研究中,我们将探索裙带菜种质的无菌处理、保存及培养技术,在此基础上研究附生微生物对种质资源保存、培养及利用的影响,提升海带目褐藻种质保存的技术水平。
ASHEN J B, GOFF L J. Molecular and ecological evidence for species specificity and coevolution in a group of marine algal-bacterial symbioses. Applied and Environmental Microbiology, 2000, 66(7): 3024-3030 DOI:10.1128/AEM.66.7.3024-3030.2000 |
BELL W, MITCHELL R. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biological Bulletin, 1972, 143: 265-277 DOI:10.2307/1540052 |
BOLGER A M, LOHSE M, USADEL B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 2014, 30(15): 2114-2120 DOI:10.1093/bioinformatics/btu170 |
BOLYEN E, RIDEOUT J R, DILLON M R, et al. Author correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 2019, 37(8): 1091 |
Bureau of Fisheries, Ministry of Agriculture and Rural Affairs, National Fisheries Technology Extension Center, China Society of Fisheries. China fishery statistical yearbook. Beijing: China Agriculture Press, 2020 [农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会. 2020中国渔业统计年鉴. 北京: 中国农业出版社, 2020]
|
CHE Y L, WANG H, HU H Y, et al. Research progresses on analytical technologies used in microbial community structure and diversity. Ecology and Environment, 2005, 14(1): 127-133 [车玉伶, 王慧, 胡洪营, 等. 微生物群落结构和多样性解析技术研究进展. 生态环境, 2005, 14(1): 127-133 DOI:10.3969/j.issn.1674-5906.2005.01.027] |
CHEN D, LIN G H, SHEN S Z. Studies on alginic acid decomposing bacteria Ⅱ. Rot disease of Laminaria summer sporelings caused by alginic acid decomposing bacteria. Oceanologia et Limnologia Sinica, 1981(2): 133-137 [陈  , 林光恒, 沈世泽. 褐藻酸降解菌的研究Ⅱ. 海带夏苗培育中褐藻酸降解菌与烂苗的关系. 海洋与湖沼, 1981(2): 133-137] , 林光恒, 沈世泽. 褐藻酸降解菌的研究Ⅱ. 海带夏苗培育中褐藻酸降解菌与烂苗的关系. 海洋与湖沼, 1981(2): 133-137] |
EDGAR R C, HAAS B J, CLEMENTE J C, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 2011, 27(16): 2194-2200 DOI:10.1093/bioinformatics/btr381 |
EDGAR R C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 2013, 10: 996-998 DOI:10.1038/nmeth.2604 |
EGAN S, FERNANDES N D, KUMAR V, et al. Bacterial pathogens, virulence mechanism and host defence in marine macroalgae. Environmental Microbiology, 2014, 16(4): 925-938 DOI:10.1111/1462-2920.12288 |
FUKUI Y, ABE M, KOBAYASHI M, et al. Isolation of Hyphomonas strains that induce normal morphogenesis in protoplasts of the marine red alga Pyropia yezoensis. Microbial Ecology, 2014, 68(3): 556-566 DOI:10.1007/s00248-014-0423-4 |
GAI S S, ZHANG W D. Microbiota of shell-boring conchocelis of Pyropia yezoensis determined by the next-generation sequencing. Oceanologia et Limnologia Sinica, 2016, 47(5): 990-996 [盖珊珊, 张伟东. 基于第二代测序技术的条斑紫菜(Pyropia yezoensis)贝壳丝状体附生菌群研究. 海洋与湖沼, 2016, 47(5): 990-996] |
GRICE E A, KONG H H, CONLAN S, et al. Topographical and temporal diversity of the human skin microbiome. Science, 2009, 324(5931): 1190-1192 DOI:10.1126/science.1171700 |
HONG C, SI Y X, XING Y, et al. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environmental Science and Pollution Research International, 2015, 22(14): 10788-10799 DOI:10.1007/s11356-015-4186-3 |
JIANG J Y, MA Y X, ZHANG Z Y, et al. The histopathological study on "green decay diseases" of Undaria pinnatifida in Dalian. Journal of Dalian Ocean University, 1997, 12(3): 7-12 [姜静颖, 马悦欣, 张泽宇, 等. 大连地区裙带菜"绿烂病"组织病理的研究. 大连水产学院学报, 1997, 12(3): 7-12] |
JIANG J, YANG B L, YUAN Q, et al. Isolation and identification of marine symbiotic and epiphyte microorganisms with antimicrobial activity. Chinese Journal of Marine Drugs, 2005, 24(3): 39-42 [姜健, 杨宝灵, 元起, 等. 海洋共附生微生物的分离和抗菌活性鉴定. 中国海洋药物杂志, 2005, 24(3): 39-42] |
KESHTACHER-LIEBSO E, HADAR Y, CHEN Y. Oligotrophic bacteria enhance algal growth under iron-deficient conditions. Applied and Environmental Microbiology, 1995, 61(6): 2439-2441 DOI:10.1128/aem.61.6.2439-2441.1995 |
LI J, MOU Z J, YANG H C, et al. Isolation and identification the pathogen of Pyropia yezoensis green spot disease. Progress in Fishery Sciences, 2019, 40(4): 140-146 [李杰, 牟宗娟, 杨慧超, 等. 条斑紫菜(Pyropia yezoensis)绿斑病病原菌的分离鉴定. 渔业科学进展, 2019, 40(4): 140-146] |
LI J, PANG S J, SHAN T F, et al. Changes of microbial community structures associated with seedlings of Saccharina japonica at early stage of outbreak of green rotten disease. Journal of Applied Phycology, 2020, 32(2): 1323-1327 DOI:10.1007/s10811-019-01975-7 |
LIANG S S. Analysis on the community structure of phycosphere bacteria in the filaments of Porphyra haitanensis and gametophytes of Saccharina japonica. Master´s Thesis of Fujian Normal University, 2019 [梁珊珊. 坛紫菜丝状体和海带配子体的藻际细菌群落结构分析. 福建师范大学硕士研究生学位论文, 2019]
|
MA J H. A preliminary study on the red rot disease of Porphyra yezoensis. Journal of Shanghai Fisheries University, 1996a, 5(1): 1-7 [马家海. 条斑紫菜赤腐病的初步研究. 上海水产大学学报, 1996a, 5(1): 1-7] |
MA J H. An investigation of chytrid-disease in Porphyra yezoensis in the coastal water of south Jiangsu. Journal of Shanghai Fisheries University, 1992, 1(3/4): 185-188 [马家海. 江苏省南部沿海条斑紫菜壶状菌病的调查研究. 上海水产大学学报, 1992, 1(3/4): 185-188] |
MA J H. Studies on the prevention and cure for diatom felt disease of Porphyra yezoensis. Journal of Shanghai Fisheries University, 1996b, 5(3): 163-169 [马家海. 条斑紫菜硅藻附着症防治研究. 上海水产大学学报, 1996b, 5(3): 163-169] |
MA Y X, ZHANG Z Y, FAN C J, et al. Study on the pathogenic bacteria of spot decay disease of Undaria pinnatifida in Dalian. Journal of Fishery Sciences of China, 1997, 4(3): 62-65 [马悦欣, 张泽宇, 范春江, 等. 大连地区裙带菜斑点烂病病原菌的研究. 中国水产科学, 1997, 4(3): 62-65 DOI:10.3321/j.issn:1005-8737.1997.03.012] |
MAGOČ T, SALZBERG S L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 2011, 27(21): 2957-2963 DOI:10.1093/bioinformatics/btr507 |
MARSHALL K, JOINT I, CALLOW M E, et al. Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microbial Ecology, 2006, 52(2): 302-310 DOI:10.1007/s00248-006-9060-x |
MARTIN M. Cutadapt removes adapter sequences from high- throughput sequencing reads. EMBnet Journal, 2011, 17(1): 10-12 DOI:10.14806/ej.17.1.200 |
MATSUO Y, SUZUKI M, KASAI H, et al. Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environmental Microbiology, 2003, 5(1): 25-35 |
PROVASOLI L. Media and prospects for the cultivation of marine algae. Cultures and collections of algae. Proceedings of US, Japan Conference, Hakone. Toyko: Japanese Society for Plant Physiology, 1968, 63–75
|
SAWABE T, MAKINO H, TATSUMI M, et al. Pseudoalteromonas bacteriolytica sp. nov., a marine bacterium that is the causative agent of red spot disease of Laminaria japonica. International Journal of Systematic Bacteriology, 1998, 48(3): 769-774 DOI:10.1099/00207713-48-3-769 |
SHEN M L, YANG R, LUO Q J, et al. Microbial diversity of Pyropia haitanensis phycosphere during cultivation. Acta Microbiologica Sinica, 2013, 53(10): 1087-1102 [沈梅丽, 杨锐, 骆其君, 等. 坛紫菜养殖周期中的藻际微生物多样性. 微生物学报, 2013, 53(10): 1087-1102] |
SINGH R P S, REDDY C R K. Seaweed-microbial interactions: Key functions of seaweed-associated bacteria. FEMS Microbiology Ecology, 2014, 88(2): 213-230 |
STOECK T, BASS D, NEBEL M, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Molecular Ecology, 2010, 19(S1): 21-31 |
SUN P H, QIAN K, LI X L, et al. Characterization of bacterial community structure on surface of kelp Saccharina japonica by high-throughput sequencing. Journal of Dalian Ocean University, 2017, 32(1): 7-12 [孙丕海, 钱坤, 李晓丽, 等. 基于高通量测序分析海带表面细菌群落结构. 大连海洋大学学报, 2017, 32(1): 7-12] |
SUN S, YUE H D, XIAO T. Method and ecology function of marine Cyanobacterium (Synechococcus). Science Technology and Engineering, 2005, 5(21): 1628–1633, 1645 [孙晟, 岳海东, 肖天. 海洋蓝细菌(聚球菌属, Synechococcus)的生态学意义及研究方法. 科学技术与工程, 2005, 5(21): 1628–1633, 1645] |
TANG L C, ZHONG C H, LIN Q, et al. Comparative study on the community structure of epiphytic bacteria in sporophyte and gametophyte stages of kelp Saccharina japonica. Progress in Fishery Sciences, 2020, 41(1): 58-65 [唐隆晨, 钟晨辉, 林琪, 等. 海带孢子体、配子体时期附生菌群落结构的比较. 渔业科学进展, 2020, 41(1): 58-65] |
WANG H B, LI X S, XIA Y M, et al. Isolation, identification and biological pathogen of yellow spot disease in conchocelis of Porphyra yezoensis. Marine Environmental Science, 2011, 30(3): 361–364, 408 [王洪斌, 李信书, 夏亚明, 等. 条斑紫菜丝状体黄斑病病原体分离鉴定及生物学特性研究. 海洋环境科学, 2011, 30(3): 361–364, 408] |
WANG L L, TANG X X, WANG M, et al. The roles played by alginic acid decomposing bacteria during the time of green decay disease of Laminaria japonica. Journal of Ocean University of Qingdao, 2003, 33(2): 245-248 [王丽丽, 唐学玺, 王蒙, 等. 褐藻酸降解菌在海带绿烂病发生中的作用. 青岛海洋大学学报, 2003, 33(2): 245-248] |
WU H Q. Epibacterial community structure of several macroalgae. Doctoral Dissertation of University of Chinese Academy of Sciences (Institute of Oceanology, Chinese Academy of Sciences), 2012 [武洪庆. 不同养殖海藻表面附着细菌多样性分析. 中国科学院大学(海洋研究所)博士研究生学位论文, 2012]
|
XU R L, MIAO S Y, YU M Y, et al. Harmful ciliates and dominant population in cultured ponds of algae. Journal of Tropic Oceanology, 2000, 19(3): 20-26 [徐润林, 苗素英, 余勉余, 等. 鲍苗硅藻饵料培养池中的危害性纤毛虫及其优势种群研究. 热带海洋, 2000, 19(3): 20-26] |
YANG H C, LI J, YAN Y W, et al. Pathogens, virulence mechanism, and disease control in economic marine macroalgae. Marine Sciences, 2019, 43(1): 101-111 [杨慧超, 李杰, 阎永伟, 等. 经济海藻病害及防控技术的研究进展. 海洋科学, 2019, 43(1): 101-111] |
ZHANG S J, CAI Z H, ZHU W S, et al. Advances in extracellular polymeric substances in phycosphere environment. Acta Microbiologica Sinica, 2020, 60(8): 1521-1533 [张圣洁, 蔡中华, 朱伟胜, 等. 藻际环境中胞外聚合物的研究进展. 微生物学报, 2020, 60(8): 1521-1533] |
ZOU D, XIAO L, YANG L Y, et al. Effects of phosphorus sources of different forms on phosphorus metabolism of Microcystis aeruginosa and adhesive Pseudomonas sp. Environmental Science, 2005, 26(3): 118-121 [邹迪, 肖琳, 杨柳燕, 等. 不同形态磷源对铜绿微囊藻与附生假单胞菌磷代谢的影响. 环境科学, 2005, 26(3): 118-121] |



