2. 中国水产科学研究院黄海水产研究所 农业农村部 海洋渔业与可持续发展重点实验室 山东 青岛 266071;
3. 海洋渔业科学与食物产出过程功能实验室 山东 青岛 266071;
4. 大连海洋大学水产与生命学院 辽宁 大连 116023
2. Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Key Laboratory of Marine Fisheries and Sustainable Development, Ministry of Agriculture and Rural Affairs, Qingdao 266071, China;
3. Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao 266071, China;
4. College of Fisheries and Life Science, Dalian Ocean University, Dalian 116023, China
实时荧光定量PCR (qRT-PCR)因具有灵敏性高、特异性强、重复性好和成本低等优点(Jaiswal et al, 2019),是进行基因表达检测分析最常用的技术手段。qRT-PCR有绝对定量或相对定量2种方法。其中,在qRT-PCR相对定量中,为了消除样本RNA质量、逆转录效率、实验操作等因素对检测结果的影响(Bustin, 2002; Maroufi et al, 2010),通常会引入内参基因来校正相对荧光定量实验数据,以保证实验结果的准确可靠(Dheda et al, 2005)。内参基因通常选用管家基因,如斑马鱼(Danio rerio)、半滑舌鳎(Cynoglossus semilaevis)基因表达分析使用的内参基因为EF-1α (McCurley et al, 2008; 马骞等, 2016、2015);布氏鲳鲹(Trachinotus blochii)、草鱼(Ctenopharyngodon idella)基因表达研究的内参基因为β-actin (李岩强等, 2020; 胡伟等, 2022);鲫鱼(Carassius auratus auratus)使用了RPL-7为内参基因(宋双双等, 2013)。常用的管家基因有构成细胞器骨架的基本组分的β-actin、TUB、RPL、β2M等基因和参与生物体基本生化代谢过程的EF-1α、GAPDH、PPIA和PP2A等基因。管家基因是维持细胞生命活动的典型基因,通常认为其表达水平受环境因素影响较小,并能在生物体几乎全部组织及各个生长阶段持续表达,但Yang等(2020)研究表明,管家基因的表达稳定性会受到物种和组织差异的影响,在不同物种中的表达稳定性并不一致。因此,针对特定物种筛选稳定表达的内参基因,有助于提高qRT-PCR结果的准确性。
黄带拟鲹隶属于硬骨鱼纲(Osteichthyes)、鲈形目(Perciformes)、鲹科(Carangidae)、拟鲹属(Pseudocaranx),是中上层长距离洄游鱼类(陈大刚等, 2015)。为了满足深远海生物资源开发利用的理论与技术需求,本团队正在系统开展黄带拟鲹分子生物学和遗传学研究(Wang et al, 2022),以黄带拟鲹为对象的适应性进化机制、骨骼肌分化及转化、线粒体能量代谢等相关研究也正在广泛开展。筛选合适的内参基因是开展黄带拟鲹分子生物学研究的基础。本研究以黄带拟鲹为研究对象,选择了硬骨鱼类qRT-PCR广泛使用的9个内参基因,对其在各组织中表达的稳定性进行评价分析,以期为黄带拟鲹功能基因的标准化和定量化检测提供稳定的内参基因,为后期全面系统开展黄带拟鲹分子生物学和遗传学研究提供技术支撑。
1 材料与方法 1.1 实验材料黄带拟鲹样品取自大连天正实业有限公司养殖场。选取健康成鱼3尾,体重为(1 488.72±135.95) g,体长为(36.27±0.63) cm,解剖并分别取其10个组织(脑、鳃、心脏、肠、肾、肝、脾、胃、慢肌和快肌)。PBS缓冲液冲洗,去除肌膜和其他结缔组织,置于无菌冻存管,立即将样品投入液氮中速冻,转移至–80 ℃超低温冰箱中保存,用于后续总RNA的提取。
1.2 方法 1.2.1 RNA提取与cDNA合成取1.1保存的3尾黄带拟鲹10个组织各30 mg,分别剪碎、匀浆,使用TaKaRa RNAiso Plus (宝生物)试剂分别提取总RNA,具体操作按照说明书进行。通过1%琼脂糖凝胶电泳检测提取RNA样品的完整性,并利用Nano300微量分光光度计(奥盛)检测所提取RNA的浓度与纯度。总RNA的OD260 nm/280 nm及OD260 nm/OD230 nm值均在1.8~2.2之间,表明所提取的RNA纯度较好。
使用TaKaRa PrimeScriptTM RT Reagent Kit with gDNA Eraser (宝生物)将提取的RNA样品反转录为cDNA,具体按照反转录操作说明书进行。将cDNA样品置于–20 ℃保存备用。
1.2.2 引物设计和标准曲线构建根据课题组的黄带拟鲹转录组数据(Wang et al, 2022),利用在线工具Primer5设计9个内参基因(β-actin、RPL13、EF-1α、GAPDH、HPRT、PPIA、β2M、TUB和PP2A) (表 1),由青岛华大基因生物科技有限公司合成。
|
|
表 1 9个内参基因及验证基因的引物序列 Tab.1 Primer sequences for the 9 reference genes and test gene |
将1.2.1得到的每尾黄带拟鲹10个组织的cDNA样品等质量混合,按1∶3比例进行7个梯度稀释(1/3、1/9、1/27、1/81、1/243、1/729、1/2187),以其为模板进行qRT-PCR,构建标准曲线,相关系数R2为0.952~0.999;扩增效率为94.4%~104.6% (表 1),符合qRT-PCR实验对引物的要求。
1.2.3 实时荧光定量PCR将1.2.1得到的cDNA稀释5倍作为模板,利用ABI 7500实时荧光定量PCR仪(Applied Biosystems, 美国)进行扩增。使用TaKaRa TB Green® Premix Ex TaqTM Ⅱ (宝生物)试剂盒配制反应液,反应体系:Premix Ex Taq Ⅱ (2×) 10 μL,ROX reference dye Ⅱ (50×) 0.4 μL,10 μmol/L上下游引物各0.8 μL,25 ng cDNA模板,用双蒸水补足至20 μL,所有操作在冰上进行。qRT-PCR扩增程序:95 ℃预变性30 s后进入循环;95 ℃变性5 s,60 ℃退火34 s,收集荧光信号,共40个循环;后运行熔解曲线分析程序:95 ℃变性15 s,60 ℃稳定60 s,以0.05 ℃/s的速度上升到95 ℃,记录荧光信号的变化,95 ℃稳定15 s,60 ℃退火15 s。每个样品设3个技术重复,并设阴性对照。
1.2.4 内参基因稳定性预测经7500 System SDS软件(Applied Biosystems, 美国)处理qRT-PCR结果,得到内参基因循环阈值(Ct)。通过公式:Cq=Ct×log(2)/ logE (其中,E是每个引物对的扩增效率)将各样品qRT-PCR得到的Ct值转换为Cq值。
根据4种不同的算法预测内参基因的表达稳定性,包括BestKeeper、NormFinder、geNorm和RefFinder模型。BestKeeper根据Ct值的变异系数(CV)和标准差(SD)评估内参基因的表达稳定性(Pfaffl et al, 2004)。对于NormFinder和geNorm,利用公式Q=2Cqmin-Cqsample计算得到Q值(Cqmin为基因在样品中的最小Cq值;Cqsample为基因在相关样品中的Cq值)。NormFinder引入一个ANOV导向模型来计算Q值组内和组间变异,并根据稳定值(SV)对内参基因进行排序(Andersen et al, 2004)。geNorm通过Q值计算内参基因相对于同一分析中包含的所有其他基因的平均成对变异来确定基因表达稳定性值M(Vandesompele et al, 2002)。综合分析工具RefFinder (https://www.heartcure.com.au/for-researchers/)根据几何平均值(GM)对内参基因的稳定性进行分析(Xie et al, 2011)。
1.2.5 内参基因稳定性验证利用qRT-PCR分析成肌分化蛋白基因myod1在黄带拟鲹红肌、白肌2个肌肉组织与肾、鳃2个非肌肉组织的表达情况,引物设计方法同1.2.2 (表 1),qRT-PCR方法参照1.2.3。在引入上述分析预测得到的稳定性较好的RPL13和EF-1α,及稳定性较差的β2M、β-actin和GAPDH分别为内参基因的情况下,对myod1在同一个组织中的相对表达水平进行差异性分析,验证所选内参基因稳定性和可靠性。
1.2.6 数据统计分析使用2–ΔΔCt方法对基因表达进行相对量化(Livak et al, 2001),用平均值±标准差(Mean±SD)表示。使用SPSS软件中的单因素方差分析(one-way ANOVA)和t检验进行差异显著性检验。
2 结果 2.1 内参基因表达丰度qRT-PCR的Ct值可直接反映基因的表达丰度,Ct值大小与该基因表达量高低呈负相关。如图 1所示,9个内参基因在黄带拟鲹所有组织的平均Ct值范围为20.85~32.92,表达丰度从高到低排序依次是RPL13 > EF-1α > β2M > β-actin > PPIA > HPRT > TUB > PP2A > GAPDH。其中,RPL13的平均Ct值最小,说明该基因的表达丰度最高;GAPDH的平均Ct值最大,说明该基因的表达丰度最低。
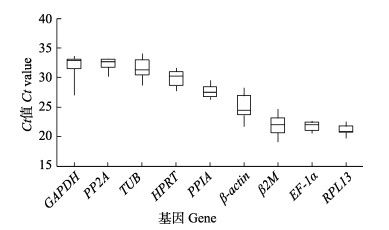
|
图 1 9个内参基因在黄带拟鲹不同组织中qRT-PCR检测的Ct值 Fig.1 Ct values of 9 reference genes in different tissues of P. dentex by qRT-PCR 箱体表示Ct值的集中范围,箱体中的横线表示中位数,箱体上下边分别表示上/下四分位数,直线上下两端分别表示最大/小值。 The box represents the concentrated range of Ct values, the horizontal line in the box represents the median, the upper and lower sides of the box respectively represent the upper/lower quartiles, and the upper and lower ends of the box respectively indicate the maximum/minimum value. |
BestKeeper通过计算标准偏差(SD)和变异系数(CV)来反映内参基因的稳定性。其中,SD值的默认阈值(图 2横线)为1.0,低于该值即认为表达稳定(Pfaffl et al, 2004);CV值反映内参基因在不同组织中的表达水平变异程度。SD值和CV值与基因的稳定性均呈负相关性。BestKeeper分析结果如图 2显示,其稳定性从高到低依次是:EF-1α (SD=0.72; CV=3.29) > RPL13 (SD=0.78; CV=3.69) > PP2A (SD=0.81; CV=2.49) > PPIA (SD=0.95; CV=3.41) > HPRT (SD=1.20; CV=4.03) > β2M (SD=1.42; CV=6.51) > TUB (SD=1.43; CV=4.51) > GAPDH (SD= 1.74; CV=5.48) > β-actin (SD=1.81; CV=7.20);有4个基因的SD值小于1.0,说明EF-1α、RPL13、PP2A和PPIA基因在黄带拟鲹10个不同组织中表达较为稳定。
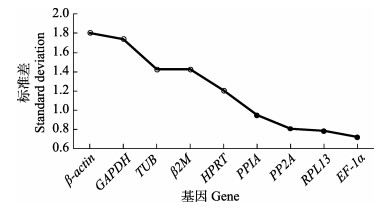
|
图 2 BestKeeper分析9个内参基因的标准差 Fig.2 Analysis of standard deviation of 9 reference genes by BestKeeper 横线代表SD值为1.0的默认阈值,实心点代表表达稳定的基因,空心点代表表达不稳定的基因。 The horizontal line indicates the BestKeeper cut-off value of 1.0, solid point indicates stable reference genes, and hollow point indicates unstable reference genes. |
NormFinder基于组间方差与组内方差计算stability value (SV),而SV值的大小与内参基因的稳定性呈负相关(Andersen et al, 2004)。NormFinder分析结果如图 3显示,9个内参基因在黄带拟鲹不同组织部位的稳定性排名从高到低依次是:RPL13 (SV=0.525) > EF-1α (SV=0.595) > HPRT (SV=0.904) > TUB (SV=0.946) > PP2A (SV=1.077) > PPIA (SV=1.284) > β2M (SV=1.775) > GAPDH (SV=2.521) > β-actin (SV=2.563)。其中,RPL13、EF1A的SV值较接近,且相比其他基因的SV值较低,说明这2个内参基因的稳定性较强。
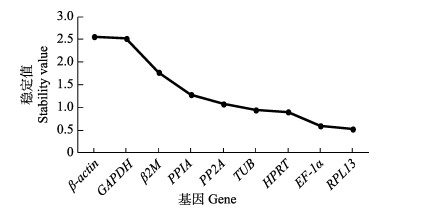
|
图 3 NormFinder分析9个内参基因的稳定值 Fig.3 Analysis of stability value of 9 reference genes by NormFinder |
根据geNorm基于单个内参基因相对于所有其他测试对照基因的成对变异进行分析。通过比较计算内参基因稳定性的M值,逐步排除最不稳定的内参基因,得到剩余内参基因的平均表达稳定性M值(Vandesompele et al, 2002)。从左边最不稳定的基因开始,根据表达稳定性的增加对基因进行排序,以右边最稳定的2个基因结束,以确定表达最稳定的内参基因。geNorm分析得到剩余内参基因的平均表达稳定性M值结果(图 4A),表明9个内参基因稳定性排名从高到低依次是:RPL13=EF-1α > β-actin > GAPDH > PP2A > TUB > PPIA > HPRT > β2M。
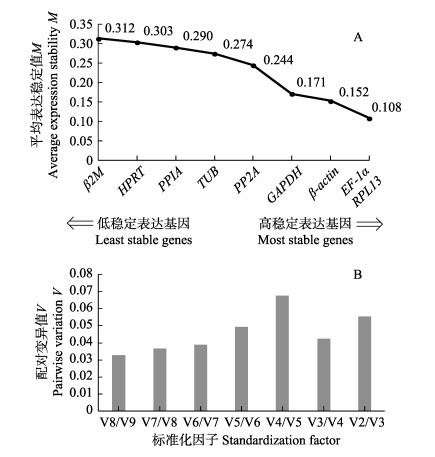
|
图 4 geNorm分析9个内参基因的平均表达稳定值(A)和配对变异值(B) Fig.4 Average expression stabilities of 9 reference genes (A) and the pairwise variation (B) analyzed by geNorm |
进一步利用geNorm分析计算配对变异值V确定所需内参基因的最佳数量。内参基因的最佳数量必须满足Vn/(n+1)低于临界值0.15。当Vn/(n+1) < 0.15,n个内参基因已达到校正目的基因表达量要求;反之,则需n+1个目的基因进行校正。图 4B结果显示,所有的成对变异系数Vn/(n+1)均小于临界值0.15,即加入更多个数的内参基因对V值影响不大,因此,2个内参基因(RPL13和EF-1α)联合使用,可达到校正目的基因表达量要求。
2.2.4 RefFinder分析RefFinder对geNorm、NormFinder、BestKeeper 3种模型进行整合分析,为单个基因分配适当的权重,综合比较和排名内参基因的表达稳定性(Xie et al, 2011)。RefFinder分析结果显示,各基因稳定性综合排名由高到低依次为RPL13 (GM=1) > EF-1α(GM=1.86) > PP2A(GM=2.71) > HPRT (GM=4.23) > PPIA(GM=4.73) > TUB(GM=6.24) > β2M (GM=6.74) > β-actin(GM=8.24) > GAPDH(GM=8.74)。其中,RPL13和EF-1α的表达稳定性最好。
2.3 内参基因稳定性验证根据4个模型对内参基因稳定性的预测结果,选择稳定性较好的RPL13和EF-1α,及稳定性较差的β2M、β-actin和GAPDH分别为内参基因,测定目标基因myod1在黄带拟鲹各组织的相对表达,并进行不同内参基因引入条件下其相对表达水平的差异性分析,以验证软件预测结果的可靠性。结果如图 5所示,在单独或联合使用RPL13和EF-1α为qRT-PCR内参基因时,myod1基因在黄带拟鲹的同一组织中的相对表达水平均无显著性差异(图 5深灰色条形柱);但以β2M、β-actin和GAPDH为内参基因时,在多数组织中其相对表达水平表现出显著性差异(图 5浅灰色条形柱)。结果表明,单独或联合使用RPL13和EF-1α作为黄带拟鲹不同组织基因表达的内参基因,能保证qRT-PCR结果的准确性和可靠性。
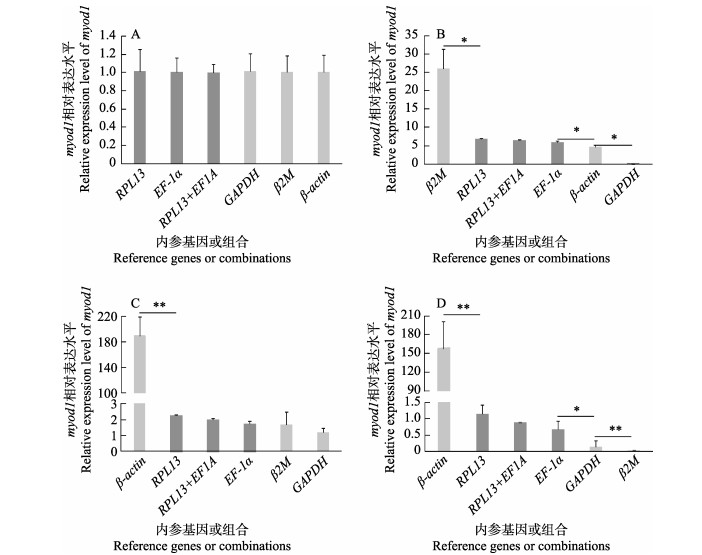
|
图 5 不同内参基因或组合分析myod1在黄带拟鲹慢肌(A)、快肌(B)、肾(C)、鳃(D) 4个组织中的相对表达水平 Fig.5 Relative expression level of myod1 in slow-twitch muscle (A), fast-twitch muscle (B), kidney (C) and gill (D) of P. dentex using different reference genes or combinations *表示差异显著(P < 0.05),**表示差异极显著(P < 0.01)。 * shows the significant difference (P < 0.05), and ** shows the highly significant difference (P < 0.01). |
当前,qRT-PCR是检测基因表达的重要方法,也是深入挖掘基因功能必不可少的实验技术。稳定的内参基因要求其表达水平不受研究条件的影响且在不同组织之间稳定表达(Zhang et al, 2005),能够准确量化qRT-PCR相对定量结果。但Yang等(2020)研究发现,有的内参基因的表达会受到不同物种、不同组织、不同发育阶段以及不同环境条件的影响。因此,并不是所有的物种和条件下都适合选用同一种管家基因,如18S rRNA是青鳉(Oryzias latipes)、脊尾白虾(Exopalaemon carinicauda)较为理想的内参基因(Zhang et al, 2007; 薛蓓等, 2017),但在菊黄东方鲀(Takifugu flavidus)、异育银鲫(Carassius auratus gibelio)、三倍体虹鳟(Oncorhynchus mykiss)、草鱼等物种中却不适用(蒋彩云等, 2020; 费越越等, 2020; 苏晓燕等, 2022; 周瑞雪等, 2009)。目前,没有发现一种不受外界因素影响、能够在所有物种和组织细胞中都能稳定表达的理想内参基因(Bruge et al, 2011),因此,在qRT-PCR实验之前,针对特定物种筛选其各组织均能稳定表达的内参基因就显得尤为重要。
3.2 内参基因的稳定性评价qRT-PCR内参基因常常选择在不同组织中具有一定保守性的管家基因,通常认为管家基因在不同的组织细胞中能够稳定表达。本研究选择9个常用的管家基因为内参基因,根据其在黄带拟鲹10个组织的表达情况,运用4种模型预测其稳定性。结果显示,4种模型预测结果的排序基本一致,即RPL13和EF-1α在黄带拟鲹不同组织的表达最具稳定性。其中,与三倍体虹鳟5个内参基因的CV值范围3.94~ 15.36 (苏晓燕等, 2022)相比,本研究BestKeeper分析9个内参基因2.49~7.20的CV值范围更低;本研究NormFinder分析9个内参基因的SV值范围0.525~2.563,与三倍体虹鳟(苏晓燕等, 2022)、达氏鲟(Acipenser dabryanus) (武梦斌等, 2020)的研究结果接近。同时,RPL13、EF-1α在黄带拟鲹不同组织的表达丰度较适中,符合理想内参基因的条件(朱芷葳等, 2006)。已有研究表明,RPL13在驼背鲈(Cromileptes altivelis)不同免疫器官和尖裸鲤(Oxygymnocypris stewarti)中被鉴定为表达最稳定的内参基因(Chen et al, 2021; 孙海成等, 2019);EF-1α是斑马鱼、黄鳝(Monopterus albus)、达氏鲟表达最稳定的内参基因(McCurley et al, 2008; Hu et al, 2014; 武梦斌等, 2020)。同时,4种模型预测到其他7个内参基因的稳定性均较差,但排序结果稍有差异,这可能是内参基因稳定性算法的不同造成的(Ward et al, 2015)。
在多内参基因表达稳定性分析方面,Bustin等(2009)建议在一些对基因表达定量准确度要求非常高的实验中选取适宜的2个基因作为内参基因。因为双内参组合能更准确地分析目的基因在不同组织的相对表达量,提高实验结果的精确度,确保目的基因相对表达结果的可靠性(苏晓燕等, 2022)。本研究也证实了联合使用RPL13和EF-1α两个内参基因,结果具有更好的稳定性和准确性。
3.3 内参基因稳定性评价结果的验证为了进一步验证模型预测结果的准确性,本研究利用myod1在黄带拟鲹慢肌、快肌、肾、鳃等各组织的相对表达进行不同内参基因引入条件下的相对表达量分析。myod1是脊椎动物胚胎期肌肉发育的主导调控基因之一,对骨骼肌的形成和分化起关键作用。以GAPDH为内参基因时myod1在快肌(图 5B)基本无表达,与单独或联合使用RPL13和EF-1α为内参基因时myod1主要在快肌中表达的结果截然相反,也显然与myod1主要在骨骼肌中表达的现象(孔祥福等, 2021)相矛盾;以β-actin为内参基因时,myod1在肾(图 5C)和鳃(图 5D)中超高表达,与myod1在非肌肉细胞中的表达受抑制的研究结果(Weintraub et al, 1991)相矛盾。该结果验证了4个模型的预测结果,表明RPL13和EF-1α稳定性确实较好,是适用于黄带拟鲹多个组织的基因表达水平研究的内参基因。本研究筛选了黄带拟鲹不同组织中稳定的内参基因,可为后期全面系统开展黄带拟鲹分子生物学和遗传学研究提供依据。
ANDERSEN C L, JENSEN J L, ØRNTOFT T F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 2004, 64(15): 5245-5250 DOI:10.1158/0008-5472.CAN-04-0496 |
BRUGE F, VENDITTI E, TIANO L, et al. Reference gene validation for qPCR on normoxia-and hypoxia-cultured human dermal fibroblasts exposed to UVA: Is β-actin a reliable normalizer for photoaging studies?. Journal of Biotechnology, 2011, 156(3): 153-162 DOI:10.1016/j.jbiotec.2011.09.018 |
BUSTIN S A, BENES V, GARSON J A, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 2009, 55(4): 611-622 DOI:10.1373/clinchem.2008.112797 |
BUSTIN S A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. Journal of Molecular Endocrinology, 2002, 29(1): 23-39 DOI:10.1677/jme.0.0290023 |
CHEN D G, ZHANG M Z. Marine fishes of China. Qingdao: China Ocean University Press, 2015: 1096-1097 [陈大刚, 张美昭. 中国海洋鱼类. 青岛: 中国海洋大学出版社, 2015: 1096-1097]
|
CHEN X, SUN Y, ZHANG P, et al. Screening of stable internal reference genes by quantitative real-time PCR in humpback grouper Cromileptes altivelis. Journal of Oceanology and Limnology, 2021, 39(5): 15 |
DHEDA K, HUGGETT J F, CHANG J S, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Analytical Biochemistry, 2005, 344(1): 141-143 DOI:10.1016/j.ab.2005.05.022 |
FEI Y Y, NAN X Y, YU L, et al. Screening of reference genes in allogynogenetic silver crucian carp Carassius auratus gibelio. Fisheries Science, 2020, 39(3): 306-315 [费越越, 南星羽, 余路, 等. 异育银鲫内参基因的筛选. 水产科学, 2020, 39(3): 306-315] |
HU Q, GUO W, GAO Y, et al. Reference gene selection for real-time RT-PCR normalization in rice field eel (Monopterus albus) during gonad development. Fish Physiology and Biochemistry, 2014, 40(6): 1721-1730 DOI:10.1007/s10695-014-9962-3 |
HU W, WU H, YUAN H W, et al. Molecular cloning and expression analysis of the melatonin receptor gene in grass carp (Ctenopharyngodon idella). Progress in Fishery Sciences, 2022, 43(1): 141-152 [胡伟, 吴慧, 袁汉文, 等. 草鱼褪黑激素受体Mtnr1基因克隆及组织表达分析. 渔业科学进展, 2022, 43(1): 141-152] |
JAISWAL P S, KAUR N, RANDHAWA G S. Identification of reference genes for qRT-PCR gene expression studies during seed development and under abiotic stresses in Cyamopsis tetragonoloba. Crop Science, 2019, 59(1): 252-265 DOI:10.2135/cropsci2018.05.0313 |
JIANG C Y, QIAO K, XU M, et al. Selection of appropriate reference genes for real-time quantitative PCR analysis in Takifugu flavidus. Journal of Fisheries Research, 2020, 42(2): 105-116 [蒋彩云, 乔琨, 许旻, 等. 利用RT-qPCR技术筛选菊黄东方鲀的最适内参基因. 渔业研究, 2020, 42(2): 105-116] |
KONG X F, HE Y, SONG W H, et al. Preliminary functional analysis of MyoD1s genes in regulating the myoblast differentiation in black rockfish (Sebastes schlegelii). Periodical of Ocean University of China (Natural Science), 2021, 51(2): 53-62 [孔祥福, 贺艳, 宋伟豪, 等. 许氏平鲉MyoD1s基因的鉴定及其调控成肌细胞分化功能的初步研究. 中国海洋大学学报(自然科学版), 2021, 51(2): 53-62] |
LI Y Q, GUO C H, YE H Z, et al. Cloning and tissue expression analysis of VEGFA gene in Trachinotus blochii. Genomics and Applied Biology, 2020, 39(3): 1020-1027 [李岩强, 郭辰浩, 叶恒振, 等. 布氏鲳鲹VEGFA基因克隆与组织表达分析. 基因组学与应用生物学, 2020, 39(3): 1020-1027] |
LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods, 2001, 25(4): 402-408 DOI:10.1006/meth.2001.1262 |
MA Q, FENG W R, LIU S F, et al. Molecular cloning and expression analysis of BMP4 gene in tongue sole (Cynoglossus semilaevis). Journal of Fishery Sciences of China, 2016, 23(3): 500-512 [马骞, 冯文荣, 柳淑芳, 等. 半滑舌鳎骨形态发生蛋白4基因的克隆和表达分析. 中国水产科学, 2016, 23(3): 500-512] |
MA Q, ZHUANG Z M, FENG W R, et al. Temporal expression of transcription factors Runx2 and Osterix during early development of Cynoglossus semilaevis. Progress in Fishery Sciences, 2015, 36(5): 1-7 [马骞, 庄志猛, 冯文荣, 等. 转录因子Runx2和Osterix在半滑舌鳎(Cynoglossus semilaevis)早期发育阶段的时序性表达分析. 渔业科学进展, 2015, 36(5): 1-7] |
MAROUFI A, VAN BOCKSTAELE E, DE LOOSE M. Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Molecular Biology, 2010, 11(1): 15-26 DOI:10.1186/1471-2199-11-15 |
MCCURLEY A T, CALLARD G V. Characterization of housekeeping genes in zebrafish: Male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Molecular Biology, 2008, 9(1): 102 DOI:10.1186/1471-2199-9-102 |
PFAFFL M W, TICHOPAD A, PRGOMET C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper- Excel-based tool using pair-wise correlations. Biotechnology Letters, 2004, 26(6): 509-515 DOI:10.1023/B:BILE.0000019559.84305.47 |
SONG S S, AN L H, ZHENG B H, et al. Vitellogenin mRNA expression in wild crucian carp (Carassius auratus auratus) from Hun River. Asian Journal of Ecotoxicology, 2013, 8(1): 121-129 [宋双双, 安立会, 郑丙辉, 等. 浑河流域野生鲫鱼卵黄蛋白原基因表达. 生态毒理学报, 2013, 8(1): 121-129] |
SU X Y, HAN B Y, MENG Y Q, et al. Stability of reference genes in different tissues of triploid rainbow trout Oncorhynchus mykiss by quantitative real-time PCR. Fisheries Science, 2022, 41(1): 35-43 [苏晓燕, 韩步鹰, 孟玉琼, 等. 三倍体虹鳟实时荧光定量PCR内参基因稳定性分析. 水产科学, 2022, 41(1): 35-43] |
SUN H C, LU X N, TONG G X, et al. Screening of reference genes for real-time quantitative PCR in Stewartxs naked high-Asian-carp Oxygymnocypris stewarti. Journal of Dalian Ocean University, 2019, 34(3): 370-375 [孙海成, 吕晓楠, 佟广香, 等. 尖裸鲤实时荧光定量PCR内参基因的筛选. 大连海洋大学学报, 2019, 34(3): 370-375] |
VANDESOMPELE J, DE PRETER K, PATTYN F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 2002, 3(7): H34 |
WANG H, LI B, YANG L, et al. Expression profiles and transcript properties of fast-twitch and slow-twitch muscles in a deep-sea highly migratory fish, Pseudocaranx dentex. PeerJ, 2022, 10: e12720 |
WARD D S, JUTTA D W, ROSWITHA W, et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS One, 2015, 10(3): e122515 |
WEINTRAUB H, DAVIS R, TAPSCOTT S, et al. The myoD gene family: Nodal point during specification of the muscle cell lineage. Science, 1991, 251(4995): 761-766 |
WU M B, YE H, YUE H M, et al. Identification of the reference genes for qRT-PCR in Dabryxs sturgeon, Acipenser dabryanus. Journal of Fishery Sciences of China, 2020, 27(7): 759-770 [武梦斌, 叶欢, 岳华梅, 等. 达氏鲟实时荧光定量PCR内参基因的筛选. 中国水产科学, 2020, 27(7): 759-770] |
XIE F L, SUN G L, STILLER J W, et al. Genome-wide functional analysis of the cotton transcriptome by creating an integrated EST database. PLoS One, 2011, 6(11): e26980 |
XUE B, ZHANG P, LI Z H, et al. Cloning, expression and stability analysis of the reference gene glyceraldehyde-3- phosphate dehydrogenase (GAPDH) in Exopalaemon carinicauda. Journal of Fishery Sciences of China, 2017, 24(5): 1003-1012 [薛蓓, 张培, 李志辉, 等. 脊尾白虾GAPDH基因的克隆及其内参基因稳定性分析. 中国水产科学, 2017, 24(5): 1003-1012] |
YANG C L, YUAN X Y, ZHANG J, et al. Comprehensive transcriptome analysis of reference genes for fruit development of Euscaphis konishii. PeerJ, 2020, 8: e8474 |
ZHANG X, DING L, SANDFORD A J. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Molecular Biology, 2005, 6(1): 4 |
ZHANG Z B, HU J Y. Development and validation of endogenous reference genes for expression profiling of medaka (Oryzias latipes) exposed to endocrine disrupting chemicals by quantitative real-time RT-PCR. Toxicological Sciences, 2007, 95(2): 356-368 |
ZHOU R X, MENG T, ZHAO F L, et al. Comparative research on the stability of internal control genes based on mRNA expression analysis of MYH gene in the grass carp, Ctenopharyngodon idellus. Genomics and Applied Biology, 2009, 28(5): 896-900 [周瑞雪, 蒙涛, 赵发兰, 等. 草鱼MYH mRNA表达量分析中采用的内参基因稳定性比较. 基因组学与应用生物学, 2009, 28(5): 896-900] |
ZHU Z R, DONG C S. The stability comparison of housekeeping genes as internal standards. Letters in Biotechnology, 2006, 17(5): 807-809 [朱芷葳, 董常生. 持家基因作为相对定量内标物的稳定性比较. 生物技术通讯, 2006, 17(5): 807-809] |



