2. 广西壮族自治区兽医研究所 广西兽医生物技术重点实验室 广西 南宁 530001
2. Guangxi Key Laboratory of Veterinary Biotechnology, Guangxi Veterinary Research Institute, Nanning 530001, China
全州禾花鲤(Cyprinus carpio var. Quanzhounensis)原产于广西桂林市全州县,体色为乌褐色,鳃盖、腹部皮肤具有半透明特点,是当地稻渔综合种养产业中重要的养殖品种之一(Peng et al, 2019)。通过与建鲤(Cyprinus carpio var. Jian)皮肤的对比研究,发现其体表缺乏可反光的鸟嘌呤结晶,且黑色素含量显著高于建鲤,初步认为是导致该群体体色差异的直接原因(武霞等, 2022)。在市场上,全州禾花鲤以自身特殊的体色表型为判断种质来源和界定价格的直接依据,因此,其鸟嘌呤结晶缺失性状成为重要经济性状。虹彩细胞是鱼类色素细胞种类之一,细胞内含有规律排列的鸟嘌呤结晶,是决定鱼体表呈现金属光泽的关键物质基础(Eskova et al, 2020)。在青鳉(Oryzias latipes) (Kimura et al, 2017)、斑马鱼(Danio rerio) (Shunya et al, 2018)中,鸟嘌呤结晶缺失性状被认为是虹彩细胞分化缺失的表现,利用上述突变材料已挖掘了pnp4a、Gbx2、sox10、tfec等虹彩细胞相关基因,逐渐揭示了调控虹彩细胞形成的分化机制(Petratou et al, 2018)。鲤具有丰富的体色遗传变异类型,是研究鱼类体色决定机制的良好材料,目前已验证了基因ASIP和MC1R在瓯江彩鲤(Cyprinus carpio var. color)黑色素的聚集与分布、黑斑形成过程中的作用。全州禾花鲤的鸟嘌呤结晶缺失性状可能负载着与鸟嘌呤结晶形成或虹彩细胞分化有关的调控机制多样性和基因突变位点(武霞等, 2022),深入解析全州禾花鲤半透明的体色性状,有望挖掘相关功能基因和阐明鲤鱼的体色决定机制。另外,全州禾花鲤作为稻田养殖品种,其生活环境与池塘、网箱养殖品种差异显著,对其抗病、抗逆和耐运输性能要求较高。鱼类皮肤作为暴露在体表的第一道保护屏障,其抗炎、损伤修复等能力对鱼体抗逆性具有重要影响(Yun et al, 2021)。目前,全州禾花鲤皮肤鸟嘌呤结晶的缺失是否导致其基础生理状态的改变尚不清楚。转录组分析是了解基因型与表型之间关系的关键途径之一,其动态性和复杂性调节生物生长、发育的各个方面(Zhao et al, 2019)。ONT (Oxford Nanopore Technologies)测序技术是基于纳米孔的新一代单分子实时电信号测序技术(Lu et al, 2016),通过直接测定全长序列,从而准确识别复杂的转录本结构,发现可变剪接、多聚腺苷酸化、融合基因,以及进行转录本表达定量,已广泛引用于动植物转录组学研究(Hou et al, 2021)。为了揭示全州禾花鲤皮肤鸟嘌呤结晶缺失性状的结构基础和转录组特征,本研究以建鲤为对照,选择鸟嘌呤结晶分布差异最显著的鳞片下层组织为研究材料,首先采用透射电镜观察组织结构,其次进行ONT全长转录组测序分析,进而了解全州禾花鲤皮肤组织鸟嘌呤结晶缺失后导致的组织结构和转录组特征差异,为全州禾花鲤体色性状解析、经济性状鉴定和种质资源利用积累资料。
1 材料与方法 1.1 实验材料全州禾花鲤(HQ)和建鲤(JH)取自广西农业良种南繁基地,2种实验鱼采用人工催产方式获得,产卵、孵化时间一致,并在同一池塘中培育,在生长至128日龄时采集样本。实验鱼经MS-222麻醉后,用镊子拔取鱼体背部鳞片,放入4 ℃ RNA保存液中,4 h后用手术刀刮取鳞片下层组织(图 1的j、h区域),–80 ℃保存,用于转录组测序;另取一份鳞片常温保存于2.5%戊二醛溶液中,用于切片和透射电镜观察。

|
图 1 全州禾花鲤和建鲤外观与鳞片 Fig.1 Appearance and scale of Cyprinus carpio var. Quanzhounensis and Cyprinus carpio var. Jian JH:建鲤;HQ:全州禾花鲤。j、h:取样本部位 JH: Cyprinus carpio var. Jian; HQ: Cyprinus carpio var. Quanzhounensis. j and h: Sample location |
取戊二醛预固定的鳞片下层组织,经0.1 mol/L磷酸钠缓冲液冲洗后,用1%锇酸固定,再用0.1 mol/L磷酸钠缓冲液冲洗,经浓度逐级递升丙酮脱水、环氧树脂浸透包埋、修块、超薄切片和自然干燥后进行醋酸铀和柠檬酸铅染色,双蒸水清洗、自然干燥,用电子JEM-1230透射电镜(日本)观察。
1.3 RNA提取、文库构建将鳞片下层组织置于液氮预冷的研钵中,研磨至粉末状之后使用RNA purification reagent (Invitrogen)提取总RNA,使用RNase-free DNase (Qiagen)去除残留DNA,使用1%琼脂糖凝胶电泳、NanoDrop分光光度计评估提取的RNA的浓度、纯度和完整性。采用cDNA-PCR测序试剂盒(SQK-PCS109)制备cDNA文库,然后用MinION Mk1B测序仪对文库进行测序。
1.4 测序数据处理与结构分析测序原始数据过滤短片段、低质量reads后,与数据库比对过滤核糖体RNA,之后通过识别reads两端引物判断全长序列。全长序列用minimap2软件(Li, 2018)与鲤参考基因组(GCF_000951615.1)比对,通过比对信息进行聚类后,使用pinfish软件得到一致性序列,使用Astalavista软件分析每个样品存在的可变剪接(AS)类型(Mazin et al, 2021),采用TAPIS pipeline识别可变多聚腺苷酸化(APA)(Abdel-Ghany et al, 2016),采用Trans Decoder (v 3.0.0)软件预测新基因潜在编码区序列(CDS) (Haas et al, 2013),使用数据库animal TFDB 3.0鉴定动物转录因子(Hu et al, 2019),使用iTAK软件预测植物转录因子(Zheng et al, 2016),采用CPC (Kong et al, 2016)、CNCI (Sun et al, 2013)、CPAT (Wang et al, 2013)、Pfam (Finn et al, 2013) 4种方法对新发现的转录本进行lncRNA预测,取其交集作为最终预测结果。
1.5 转录本差异表达分析采用Benjamini-Hochberg校正方法对P值进行校正,以每百万计数(counts per million, CPM)作为衡量转录本表达水平的指标(Cingolani et al, 2012)。使用DESeq2 R软件包对转录本进行差异表达分析,筛选标准为FDR<0.01和Fold Change≥2;最后使用KOBAS软件和GO seq R软件包对差异转录本进行KEGG分析和GO分析。
1.6 差异转录本qRT-PCR验证随机选取8个显著差异转录本进行实时荧光定量PCR (qRT-PCR)实验,验证全长转录组测序数据的准确性。用Primer 6.0软件设计转录本引物(表 1),使用Aidlab反转录试剂盒合成cDNA第一链,具体反应体系:总RNA 500 ng,5×RT Reaction mix 4 µL,Random primer 1 µL,TURE script H– RTase/RI mix 1 µL,加RNase free ddH2O至20 µL,42 ℃ 40 min,65 ℃ 10 min,反应结束后得到cDNA。qRT-PCR体系:2×SYBR® Green Supermix 5 μL,10 nmol/L正向、反向引物各0.5 μL,cDNA 1 μL,ddH2O 3 μL。反应程序:预变性95 ℃ 3 min,变性95 ℃ 10 s,延伸60 ℃ 30 s,共39个循环。最后,以eef1a基因为内参基因,利用2–ΔΔCt计算各样品的转录本相对表达量。
|
|
表 1 qRT-PCR引物序列 Tab.1 Primer sequence for qRT-PCR |
鳞片下层组织切片的透射电镜观察发现(图 2),全州禾花鲤与建鲤的组织结构在两个方面存在显著差异。首先,全州禾花鲤的组织中缺乏层叠排列的鸟嘌呤结晶体,而建鲤的组织中鸟嘌呤结晶体广泛存在,在切片中观察到鸟嘌呤结晶体脱落后形成的层叠排列腔隙。其次,全州禾花鲤组织中黑色素颗粒的数量和密度明显高于建鲤,表现为颗粒较小、深色、数量较多,这与全州禾花鲤体表颜色较深、缺少银色反光物质的现象一致。

|
图 2 电镜下的鳞片下层组织结构 Fig.2 Morphology and structure of the underlying tissue of the scale seen by electron microscope A~C:全州禾花鲤;D~F:建鲤。a:黑色素颗粒;b:堆叠鸟嘌呤结晶脱落后形成的腔隙 A~C: Cyprinus carpio var. Quanzhounensis; D~F: Cyprinus carpio var. Jian. a: Melanin granules; b: The cavity formed after the crystals of stacked guanine fall off |
对测序原始数据进行短片段和低质量序列过滤后,各样本获得高质量数据2.88~3.26 Gb,N50长度为1 159~1 428,序列数目为2 685 383~3 053 722条,平均质量值均为Q10,表明测序数据质量较好。所有样品数据过滤核糖体RNA后识别为全长序列的数目有2 203 826~2 412 500条,各样本全长序列的比例为87.06%~88.57%。将全长序列与鲤鱼基因组比对,比对率为90.35%~92.46% (表 2),符合实验要求,可用于后续结果分析。
|
|
表 2 高质量数据和全长序列统计 Tab.2 Clean reads table and statistics of full length sequence |
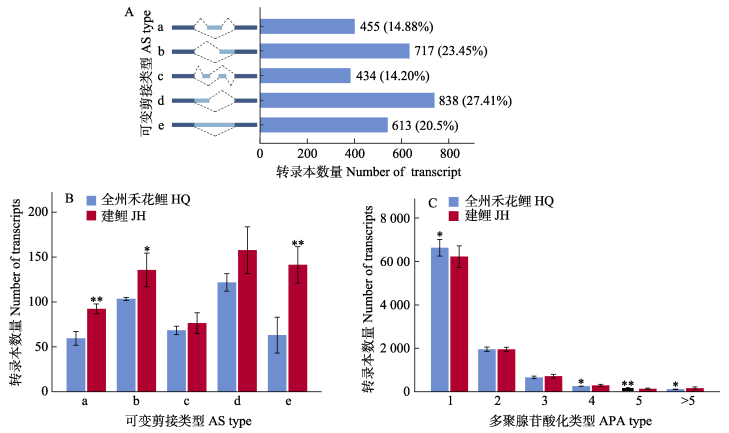
对转录本中发生的可变剪接事件进行统计,共检测出3 075个可变剪接事件,其中,外显子跳跃占14.88% (455),可变5′剪接位点占27.41% (838),可变外显子占14.20% (434),可变3′剪接位点占23.45% (713),内含子保留占20.05% (613) (图 3A)。2组之间的可变剪接数量分析显示,HQ组和JH组之间的外显子跳跃、内含子保留数量差异极显著(P<0.01),可变3'剪接位点数量差异显著(P<0.05),可变外显子、可变5′剪接位点数量差异不显著(P > 0.05) (图 3B);共检测出57 624个可变多聚腺苷酸化,HQ组和JH组中转录本3′末端有1个多聚腺苷酸化位点的转录本数分别占68.25%和65.59%,2个位点占比分别为20.17%和20.57%,3个位点占比分别为6.81%和7.57%,3个以上位点的转录本数量仅占4.82%和6.27%。其中,有5个多聚腺苷酸化位点的转录本数在组间差异极显著(P<0.01),有1个、4个和多于5个多聚腺苷酸化位点的转录本数差异显著(P<0.05),其余无差异(图 3C)。
|
图 3 可变剪接(AS)和可变多聚腺苷酸化(APA) Fig.3 Alternative splicing (AS) and variable polyadenylation (APA) a:外显子跳跃;b:可变3′剪接位点;c:可变外显子;d:可变5′剪接位点;e:内含子保留 a: Exon retention; b: Alternative 3′ splice site; c: Mutually exclusive exon; d: Alternative 5′ splice site; e: Mutually exclusive intron |
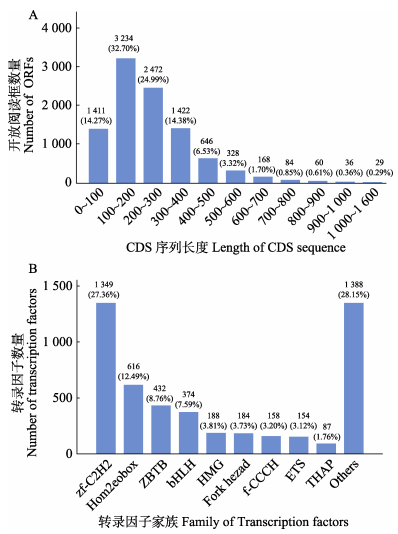
本研究共预测15 615条开放阅读框(ORF),包括9 890条完整的ORF。根据预测得到的完整ORF区,这些潜在编码区序列(CDS)中,编码100~200个氨基酸的CDS数目最多(3 234,32.70%),其次是编码200~300个氨基酸的CDS序列(2 472,24.99%)。0~100、300~400个氨基酸的CDS序列数量接近,分别占14.27%和14.38%,400个氨基酸以上的CDS序列仅占总数的13.66% (图 4A)。另外,根据新转录本预测转录因子4 930个,隶属于62个转录因子家族。其中,数量最多的3个转录因子家族分别是zf-C2H2、Homeobox和ZBTB家族,其数量分别占总数的27.36% (1349)、12.49% (616)和8.76% (432)(图 4B)。
|
图 4 预测的潜在编码区序列和转录因子 Fig.4 Predicted CDs encoded amino acid length distribution and transcription factor family |
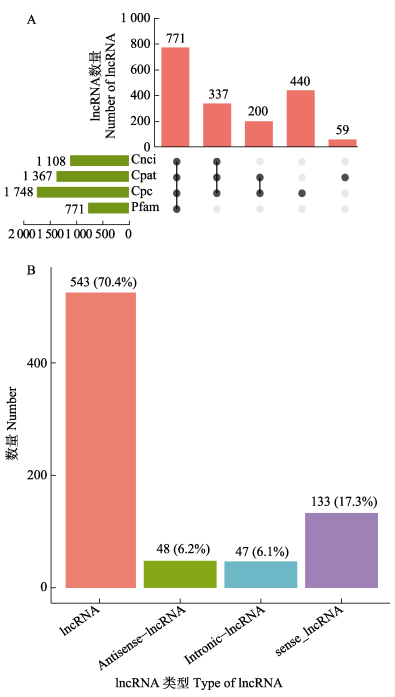
分别应用CPC、CNCI、CPAT和Pfam预测lncRNA,共预测到771个lncRNA (图 5A)。根据基因组注释位置分为4类,其中,基因间lncRNA占70.4% (543)、正义lncRNA (sense-lncRNA)占17.3% (133)、反义型lncRNA (antisense-lncRNA)占6.2% (48)、内含子型lncRNA (intronic-lncRNA)占6.1% (47) (图 5B)。
|
图 5 筛选的lncRNA及类型 Fig.5 Screened lncRNA and types |
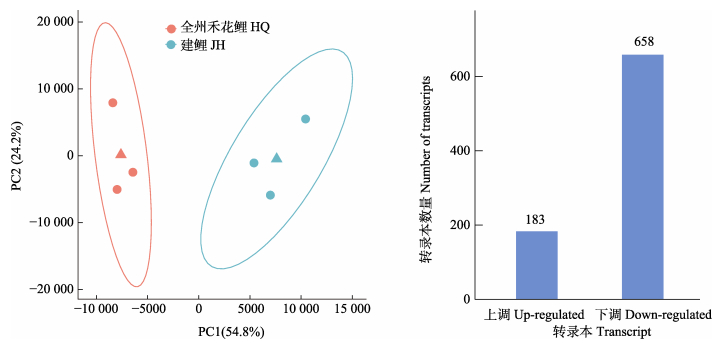
利用转录本信息开展主成分分析,全州禾花鲤(HQ)与建鲤(JH)样本分为2个类群(图 6A)。以Fold change≥2且FDR<0.01作为筛选差异表达转录本的标准,共筛选出841个差异表达转录本,与建鲤相比,在全州禾花鲤中有183个转录本上调,658个转录本下调(图 6B)。
|
图 6 样本主成分分析和差异表达转录本 Fig.6 Sample principal component analysis and differentially expressed transcripts |
差异表达转录本共有371条注释到KEGG通路,差异表达转录本主要富集在细胞外基质(ECM)–受体相互作用、粘着斑、丙氨酸–天冬氨酸–谷氨酸代谢、精氨酸生物合成、氮代谢等通路(图 7A)。网络分析显示,细胞外基质–受体相互作用和粘着斑通路高度关联,富集的转录本多数表达下调,其编码的蛋白主要有Collagen、Laminin、Chad、THBS、ITGA6和ITGB4 (图 7B),其余3个与氨基酸代谢有关的信号通路均高度关联(图 9A)。

|
图 7 差异表达转录本KEGG富集结果 Fig.7 KEGG enrichment results of differentially expressed transcripts |
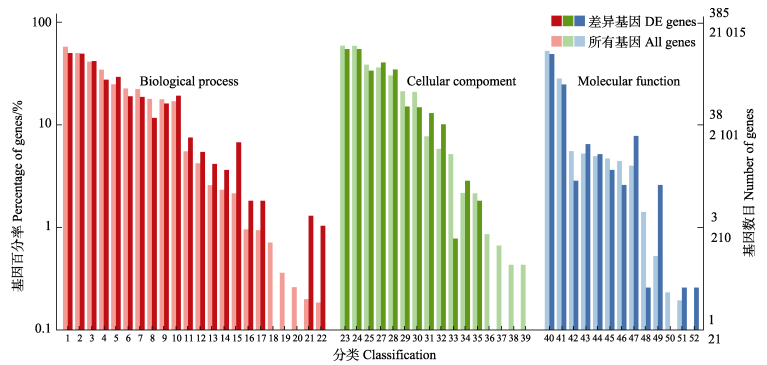
|
图 8 全州禾花鲤与建鲤鳞片下层组织的差异表达转录本GO分类统计 Fig.8 Differential expression transcripts GO classification and statistics of scaly sublayer of Cyprinus carpio var. Quanzhounensis and Cyprinus carpio var. Jian 1:细胞过程;2:单生物过程;3:生物调节;4:代谢过程;5:应激反应;6:发展进程;7:多细胞生物过程;8:细胞组成部分或生物合成;9:定位;10:信号;11:免疫系统过程;12:运动;13:多生物过程;14:生长;15:生物附着;16:生殖;17:生殖过程;18:行为;19:涉及化学突触传递的突触前过程;20:解毒;21:节奏过程;22:细胞杀伤;23:细胞;24:细胞组分;25:细胞器;26:膜;27:膜部分;28:大分子复合体;29:细胞器组分;30:细胞外区域;31:细胞外区域部分;32:膜封闭的内腔;33:细胞连接;34:超分子复合物;35:突触;36:突触部分;37:病毒粒子;38:病毒粒子部分;39:结合;40:催化活性;41:信号传感器活动;42:运输活动;43:分子功能调节剂;44:结构分子活性;45:分子传感器活动;46:核酸结合转录因子活性;47:转录因子活性,蛋白质结合;48:电子载体活动;49:抗氧化活性;50:蛋白质标签;51:化学引诱物活动;52:化学排斥物活动。 1: Cellular process; 2: Single-organism process; 3: Biological regulation; 4: Metabolic process; 5: Response to stimulus; 6: Developmental process; 7: Multicellular organismal process; 8: Cellular component organization or biogenesis; 9: Localization; 10: Signaling; 11: Immune system process; 12: Locomotion; 13: Multi-organism process; 14: Growth; 15: Biological adhesion; 16: Reproduction; 17: Reproductive process; 18: Behavior; 19: Presynaptic process involved in chemical synaptic transmission; 20: Detoxification; 21: Rhythmic process; 22: Cell killing; 23: Cell; 24: Cell part; 25: Organelle; 26: Membrane; 27: Membrane part; 28: Macromolecular complex; 29: Organelle part; 30: Extracellular region; 31: Extracellular region part; 32: Membrane- enclosed lumen; 33: Cell junction; 34: Supramolecular complex; 35: Synapse; 36: Synapse part; 37: Virion; 38: Virion part; 39: Binding; 40: Catalytic activity; 41: Signal transducer activity; 42: Transporter activity; 43: Molecular function regulator; 44: Structural molecule activity; 45: Molecular transducer activity; 46: Nucleic acid binding transcription factor activity; 47: Transcription factor activity, protein binding; 48: Electron carrier activity; 49: Antioxidant activity; 50: Protein tag; 51: Chemoattractant activity; 52: Chemorepellent activity. |
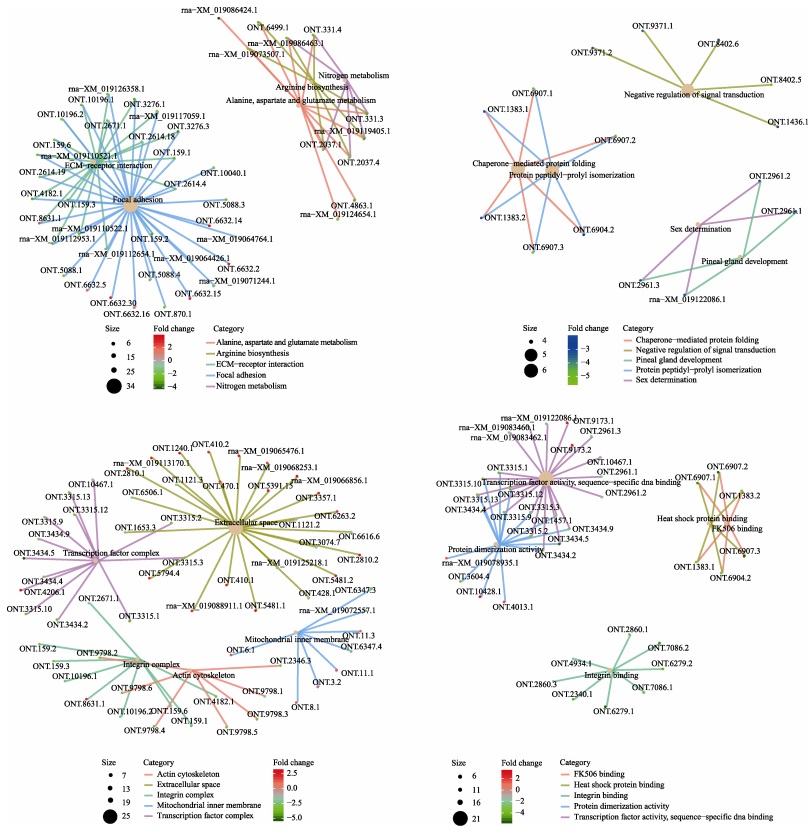
|
图 9 KEGG和GO富集网络分析 Fig.9 KEGG and GO enrichment network analysis |
对所有差异表达转录本进行GO分类注释,结果显示,在生物学过程大类下富集在22个二级分类,在细胞组分大类下富集在16个二级分类,在分子功能大类下富集在13个二级分类(图 8)。分别对GO富集显著性排在前5的Term进行网络分析显示,在生物学过程分类中,富集的转录本下调为主,主要表现为蛋白质结构相关的转录本下调(图 9B)。细胞组分分类中,富集在细胞外隙和线粒体内膜的差异表达转录本以上调为主,分别占68.00%和62.50%,富集在转录因子复合物、肌动蛋白细胞骨架和整合素复合物的差异表达转录本下调为主,分别占92.31%、100%和88.89% (图 9C)。分子功能分类中,富集的转录本下调为主,转录因子活性、序列特异性DNA结合和蛋白质二聚化活性中下调比例为90.47%和81.25%,其余3个Term中100%下调(图 9D)。对富集在细胞外隙词条的转录本进行Pfam蛋白质结构域功能注释表明,其编码的蛋白质结构域包括肿瘤坏死因子、胰蛋白酶、内分泌/趋化因子、白细胞介素等(表 3)。
|
|
表 3 富集在细胞外隙词条的转录本的Pfam注释 Tab.3 Pfam annotation of transcripts enriched in extracellular space entries |
为了验证转录组数据的准确性,随机选取差异转录本设计qRT-PCR引物对全州禾花鲤、建鲤的鳞片下层组织样品定量分析。结果显示,转录本表达定量结果与RNA-seq结果在差异倍数上存在差异,但转录本上下调趋势一致(图 10),说明利用全长转录组数据获得的转录本定量结果可信度较高。
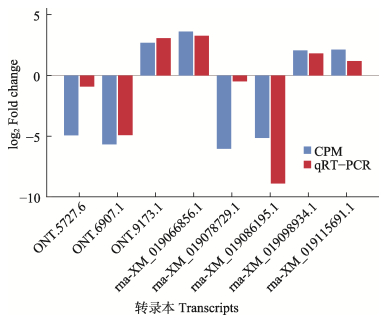
|
图 10 候选转录本qRT-PCR验证 Fig.10 qRT-PCR validation of candidate transcripts |
稻田养鱼是我国传统农业生产模式之一,具有悠久的历史和文化传承。将鲤鱼放养于稻田环境中养殖,经过长期养殖驯化、自然选择和人工选择,使其形态和体色发生不同方向的分化,产生了独特的养殖群体(Ren et al, 2018)。瓯江彩鲤(郭梁等, 2017)、全州禾花鲤(鲁翠云等, 2021)、金边鲤(何兴农, 2020)、南坡黑鲤(韦玲静等, 2020)是在体色方面最具特色的种群。其中,瓯江彩鲤体色多样,具有红、黄、白、粉、黑或混色等类型;全州禾花鲤鳃盖、腹部肌肉具有半透明特点;金边鲤背鳍两侧具有金色条带;南坡黑鲤从头至尾胸鳍基部切线往上至背部为黑色,腹部为白色或微黄色,鳍条为黑色或微黄色,构成了丰富的体色性状遗传资源。利用上述群体开展研究,解析其体色性状形成机制,对于关键功能基因挖掘和种质资源开发利用具有重要意义。近几年,对瓯江彩鲤的体色研究已发现和验证ASIP、MC1R基因在体色形成机制中的作用;对金边鲤不同颜色皮肤的转录组分析发现了ACT3、MYH2、TPMl、TPM2和TPM3基因可能在体色形成中发挥作用(Benhamed et al, 2014);对全州禾花鲤皮肤组织的前期研究已证明,鸟嘌呤结晶和黑色素含量的差异是其体色变异的物质基础(武霞等, 2022),本研究通过透射电镜观察到细胞中层叠的腔隙和黑色素颗粒增多,进一步明确了其组织显微结构差异。也体现了该群体体色与瓯江彩鲤体色变异类型的不同,为分离鉴定关键功能基因提供了性状参考;在此基础上,进行ONT测序并完成了转录组特征分析,转录本结构分析共检测出可变剪接事件3 075个,其中,可变5'剪接位点最多(27.41%),这与刀鲚(Coilia nasus)鳃组织(Gao et al, 2021)、银杏(Ginkgo biloba)叶(Ye et al, 2019)中的可变剪接类型比例明显不同,体现了其数量和比例变化的复杂性。组间比较发现,全州禾花鲤可变剪接事件的检出数量低于建鲤,其中,外显子跳跃和内含子保留数量差异为极显著水平(P<0.01),类似现象在买麻藤(Gnetum montanum)茎的不同发育阶段也有发现(Hou et al, 2021),表明建鲤皮肤组织中可变剪切调控更复杂;可变多聚腺苷酸化具有在细胞增殖和分化过程中调控基因表达的作用(Singh et al, 2018),不同亚型的相对丰度在不同的组织中差异很大(Ha et al, 2018),近期在人类胎盘细胞中的研究解释了其在B细胞向浆细胞分化过程中的调节作用(Cheng et al, 2020)。本研究中,组间可变多聚腺苷酸化也存在差异,其中,具有5个多聚腺苷酸化位点的转录本数在组间差异极显著(P<0.01),说明2种鲤鱼皮肤组织基因转录中存在不同的多聚腺苷酸化调节,但它是否参与体色形成或色素细胞分化仍不清楚。另外,本研究在新发现的转录本中共预测9 890条完整的开放阅读框、4 930个转录因子及771个lncRNA,为完善鲤鱼基因注释提供了资料,其功能作用还需进一步研究。
鱼类皮肤颜色和图案体色的形成和维持受一系列基因、细胞、生理因素和环境因素的复杂调控,且体色构成与色素合成、皮肤、鳞片所含色素细胞的种类、数量、分布及色素细胞之间的相互作用具有密切联系(Nüsslein-Volhard et al, 2017)。不同颜色的鱼类皮肤转录组研究揭示其基本差异和特征,大颚细锯脂鲤(Pristella maxillaris)银白色突变体与全透明突变体的差异表达基因在ECM–受体相互作用、蛋白质消化和吸收、蛋白酶体、糖酵解/糖异生、DNA复制和嘌呤代谢通路显著富集(Bian et al, 2019)。透明草金鱼(Carassius auratus)皮肤中差异表达基因的KEGG通路分析显示,MPKA通路、肌动蛋白细胞骨架调节通路、嘌呤代谢通路等GO分析差异基因主要富集在微管、微管结合、微管锚定等条目(刘肖莲等, 2022)。而锦鲤(Cyprinus carpio)白色皮肤和红色皮肤的差异表达基因在肌动蛋白细胞骨架调节、粘着斑通路等代谢途径富集(史东杰等, 2019)。对本研究中筛选出的841个差异转录本进行KEGG和GO分析显示,其主要在ECM–受体相互作用、粘着斑等通路显著富集,以及在细胞外隙、线粒体内膜等词条富集,与上述研究结果既有差异,也存在相同的富集通路(如:ECM–受体相互作用),可能是不同鱼类品种体色决定机制中存在的共同转录本调控信号。在大颚细锯脂鲤(Bian et al, 2019)、草金鱼(刘肖莲等, 2022)及斑马鱼(Shunya et al, 2018)透明突变体皮肤中,差异表达转录本或基因共同在嘌呤代谢相关途径富集,而本研究结果的显著不同点是未发现此类情况,这未能与组织中鸟嘌呤结晶缺失的性状特点形成直接对应,可能是全州禾花鲤该性状由细胞内鸟嘌呤结晶体形成的关键环节障碍导致,而与鸟嘌呤代谢无关,后续性状解析研究可关注鸟嘌呤在细胞内形成晶体的生物机制。
皮肤是鱼类抵御外源刺激的第一道物理屏障和非特异性免疫屏障,是沟通和接触外部因素的关键接口,其生理功能的完整性对个体生存和适应环境极为重要(Benhamed et al, 2014; Su et al, 2011; Rakers et al, 2011)。本研究对2种鲤皮肤差异表达转录本的分析结果显示,最显著富集的通路或词条从空间上均指向同一组织结构即细胞外基质(ECM)。研究表明,ECM是由结构和功能大分子组成的复杂混合物,在组织和器官的形态发生以及维持细胞和组织的结构和功能方面起着重要作用(Badylak et al, 2015)。KEGG分析显示,在最显著富集的ECM–受体相互作用通路中,转录本编码的胶原蛋白、层粘连蛋白和软骨蛋白为ECM重要组成部分,也是ECM组织力学和粘弹性的关键调节因子(Theocharis et al, 2016; Connizzo et al, 2018; Sauer et al, 2019)。研究表明,细胞外基质的粘弹性会影响基本的细胞进程,从而对组织生理功能产生重要影响(Chaudhuri et al, 2020)。在GO分析富集的细胞间隙词条中,差异表达转录本编码的肿瘤坏死因子、白细胞介素、表皮生长因子结构域、纤维蛋白原β和γ链等与非特异性免疫、损伤修复等生理过程有关。据此推测,这些作用于同一组织位置(细胞外空间)的RNA表达差异,在信号传导到的角度有可能导致细胞与细胞外基质相互作用的差异,从而影响皮肤细胞的粘附、迁移、分化、增殖和凋亡过程。也可能在结构上导致皮肤细胞外基质的组成、密度和构象变化,进而造成皮肤在抗炎、损伤修复等生理进程的变化(Poole et al, 2021)。但全州禾花鲤是否产生上述改变尚未见报道,相关工作有待开展。总之,全州禾花鲤作为稻田养殖鲤鱼群体,其面临的养殖环境和活体运输条件与其他养殖形式不同,稻田的浅水条件以及山区稻鱼种养要求养殖品种具有更高的抗逆能力。除鸟嘌呤结晶缺失作为其重要经济性状之一被研究解析之外,应关注与该性状关联的其他皮肤抗性变化,为养殖生产和种质改良提供科学依据。
ABDEL-GHANY S E, HAMILTON M, JACOBI J L, et al. A survey of the sorghum transcriptome using single-molecule long reads. Nature Communication, 2016, 7: 11706 DOI:10.1038/ncomms11706 |
BADYLAK S F, FREYTES D O, GILBERT T W. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomaterialia, 2015, 23: S17-S26 DOI:10.1016/j.actbio.2015.07.016 |
BENHAMED S, GUARDIOLA F A, MARS M, et al. Pathogen bacteria adhesion to skin mucus of fishes. Veterinary Microbiology, 2014, 171(1/2): 1-12 |
BIAN F, YANG X, OU Z, et al. Morphological characteristics and comparative transcriptome analysis of three different phenotypes of Pristella maxillaris. Frontiers in Genetics, 2019, 10: 698 DOI:10.3389/fgene.2019.00698 |
CHAUDHURI O, COOPER-WHITE J, JANMEY P A, et al. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature, 2020, 584(7822): 535-546 DOI:10.1038/s41586-020-2612-2 |
CHENG L C, ZHENG D, BALJINNYAM E, et al. Widespread transcript shortening through alternative polyadenylation in secretory cell differentiation. Nature Communication, 2020, 11(1): 3182 DOI:10.1038/s41467-020-16959-2 |
CINGOLANI P, PLATTS A, LE L W, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 2012, 6(2): 80-92 DOI:10.4161/fly.19695 |
CONNIZZO B K, GRODZINSKY A J. Multiscale poroviscoelastic compressive properties of mouse supraspinatus tendons are altered in young and aged mice. Journal of Biomechanical Engineering, 2018, 140(5): 0510021-0510028 |
ESKOVA A, FROHNHOFER H G, NUSSLEIN-VOLHARD C, et al. Galanin signaling in the brain regulates color pattern formation in zebrafish. Current Biology, 2020, 30(2): 298-303 DOI:10.1016/j.cub.2019.11.033 |
FINN R D, BATEMAN A, CLEMENTS J, et al. Pfam: The protein families database. Nucleic Acids Research, 2014, 42: D222-D230 DOI:10.1093/nar/gkt1223 |
GAO J, XU G, XU P. Gills full-length transcriptomic analysis of osmoregulatory adaptive responses to salinity stress in Coilia nasus. Ecotoxicology and Environmental Safety, 2021, 226: 112848 DOI:10.1016/j.ecoenv.2021.112848 |
GUO L, REN W Z, HU L L, et al. Morphological traits of indigenous field carps maintained in traditional rice-based farming systems. Chinese Journal of Applied Ecology, 2017, 28(2): 665-672 [郭梁, 任伟征, 胡亮亮, 等. 传统稻鱼系统中"田鲤鱼"的形态特征. 应用生态学报, 2017, 28(2): 665-672] |
HA K C H, BLENCOWE B J, MORRIS Q. QAPA: A new method for the systematic analysis of alternative polyadenylation from RNA-seq data. Genome Biology, 2018, 19(1): 45 DOI:10.1186/s13059-018-1414-4 |
HAAS B J, ALEXIE P, YASSOUR M, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nature Protocols, 2013, 8: 1494-1512 DOI:10.1038/nprot.2013.084 |
HE X N. Investigation on resources of black carp in Nanpo. Graziery Veterinary Sciences, 2020(6): 10-11 [何兴农. 南坡黑鲤资源调查. 畜牧兽医科学, 2020(6): 10-11] |
HOU C, LIAN H, CAI Y, et al. Comparative analyses of full-length transcriptomes reveal Gnetum luofuense stem developmental dynamics. Frontiers in Genetics, 2021, 12: 615284 DOI:10.3389/fgene.2021.615284 |
HU H, MIAO Y R, JIA L H, et al. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Research, 2019, 47(D1): D33-D38 DOI:10.1093/nar/gky822 |
KIMURA T, TAKEHANA Y, NARUSE K. pnp4a is the causal gene of the medaka iridophore mutant guanineless. G3 (Bethesda), 2017, 7(4): 1357-1363 DOI:10.1534/g3.117.040675 |
KONG L, ZHANG Y, YE Z Q, et al. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Research, 2016, 36: 345-349 |
LI H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics, 2018, 34(18): 3094-3100 DOI:10.1093/bioinformatics/bty191 |
LIU X L, LI C Y, BAI X H, et al. Transcriptome analysis of transparent skin in grass goldfish Carassius auratus. Fisheries Science, 2022, 41(1): 110-115 [刘肖莲, 李春艳, 白晓慧, 等. 透明草金鱼的皮肤转录组分析. 水产科学, 2022, 41(1): 110-115] |
LU C Y, DU X S, ZHENG X H, et al. Analysis of genetic structure and genetic distance between individuals of Jinbian carp by QTL markers. Freshwater Fisheries, 2021, 51(4): 58-64 [鲁翠云, 杜雪松, 郑先虎, 等. 金边鲤群体的遗传结构及个体间遗传差异的QTL标记分析. 淡水渔业, 2021, 51(4): 58-64] |
LU H Y, GIORDANO F, NING Z M. Oxford Nanopore MinION sequencing and genome assembly. Genomics, Proteomics and Bioinformatics, 2016, 14(5): 265-279 DOI:10.1016/j.gpb.2016.05.004 |
MAZIN P V, KHAITOVICH P, CARDOSO-MOREIRA M, et al. Alternative splicing during mammalian organ development. Nature Genetics, 2021, 53(6): 925-934 DOI:10.1038/s41588-021-00851-w |
NÜSSLEIN-VOLHARD C, SINGH A P. How fish color their skin: A paradigm for development and evolution of adult patterns. Bioessays, 2017, 39(3): 1600231 DOI:10.1002/bies.201600231 |
PENG J, ZENG D, HE P, et al. mRNA and microRNA transcriptomics analyses in intermuscular bones of two carp species, rice flower carp (Cyprinus carpio var. Quanzhounensis) and Jian carp (Cyprinus carpio var. Jian). Comparative Biochemistry and Physiology D Genomics and Proteomics, 2019, 30: 71-80 DOI:10.1016/j.cbd.2019.01.013 |
PETRATOU K, SUBKHANKULOVA T, LISTER J A, et al. A systems biology approach uncovers the core gene regulatory network governing iridophore fate choice from the neural crest. PLoS Genetics, 2018, 14(10): e1007402 DOI:10.1371/journal.pgen.1007402 |
POOLE J J A, MOSTACO-GUIDOLIN L B. Optical microscopy and the extracellular matrix structure: A review. Cells, 2021, 10(7): 1760 DOI:10.3390/cells10071760 |
RAKERS S, GEBERT M, UPPALAPATI S, et al. Fish matters: The relevance of fish skin biology to investigative dermatology. Experimental Dermatology, 2011, 19: 313-324 |
REN W, HU L, GUO L, et al. Preservation of the genetic diversity of a local common carp in the agricultural heritage rice-fish system. Proceedings of the National Academy of Sciences, 2018, 201709582 |
SAUER F, OSWALD L, ARIZA DE S A, et al. Collagen networks determine viscoelastic properties of connective tissues yet do not hinder diffusion of the aqueous solvent. Soft Matter, 2019, 15(14): 3055-3064 DOI:10.1039/C8SM02264J |
SHI D J, HU J Y, WANG S S, et al. Transcriptome analysis of skin of red and white koi carp Cyprinus carpio haematopterus. Journal of Dalian Fisheries University, 2019, 34(4): 475-481 [史东杰, 胡金有, 王赛赛, 等. 红白锦鲤皮肤转录组测序分析. 大连海洋大学学报, 2019, 34(4): 475-481] |
SHUNYA H, MASAKI S, JINGXIN W, et al. The N-terminal domain of gastrulation brain homeobox 2 (Gbx2) is required for iridophore specification in zebrafish. Biochemical and Biophysical Research Communications, 2018, 502(1): 104-109 DOI:10.1016/j.bbrc.2018.05.128 |
SINGH I, LEE S H, SPERLING A S, et al. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nature Communications, 2018, 9(1): 1716 DOI:10.1038/s41467-018-04112-z |
SU Y. Isolation and identification of pelteobagrin, a novel antimicrobial peptide from the skin mucus of yellow catfish (Pelteobagrus fulvidraco). Comparative Biochemistry and Physiology B Biochemistry and Molecular Biology, 2011, 158(1): 149-154 |
SUN L, LUO H, BU D, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Research, 2013, 41(17): e166 DOI:10.1093/nar/gkt646 |
THEOCHARIS A D, SKANDALIS S S, GIALELI C, et al. Extracellular matrix structure. Advanced Drug Delivery Reviews, 2016, 97: 4-27 DOI:10.1016/j.addr.2015.11.001 |
WANG L, PARK H J, DASARI S, et al. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Research, 2013, 41(6): e74 DOI:10.1093/nar/gkt006 |
WEI L J, LÜ Y J, ZOU H, et al. Skin melanin synthesis of Jinbian carp and expression analysis of transport related genes. Journal of Southern Agriculture, 2020, 51(2): 453-460 [韦玲静, 吕业坚, 邹辉, 等. 金边鲤皮肤黑色素合成及转运相关基因表达分析. 南方农业学报, 2020, 51(2): 453-460] |
WU X, XU Y L, WEN L T, et al. Composition, distribution and pigment content of chromatophores in rice flower carp in Quanzhou. Journal of Guangxi Normal University (Natural Science), 2022, 40(6): 222-229 [武霞, 许艺兰, 文露婷, 等. 全州禾花鲤色素细胞组成、分布及色素含量观测. 广西师范大学学报(自然科学版), 2022, 40(6): 222-229] |
YE J B, CHENG S Y, ZHOU X, et al. A global survey of full-length transcriptome of Ginkgo biloba reveals transcript variants involved in flavonoid biosynthesis. Industrial Crops and Products, 2019, 139: 111547 DOI:10.1016/j.indcrop.2019.111547 |
YUN T, SHIN S, BANG K, et al. Skin wound healing rate in fish depends on species and microbiota. International Journal of Molecular Sciences, 2021, 22(15): 7804 DOI:10.3390/ijms22157804 |
ZHAO L, ZHANG H, KOHNEN M V, et al. Analysis of transcriptome and epitranscriptome in plants using PacBio Iso-Seq and nanopore-based direct RNA sequencing. Frontiers in Genetics, 2019, 10: 253 DOI:10.3389/fgene.2019.00253 |
ZHENG Y, JIAO C, SUN H, et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Molecular Plant, 2016, 9(12): 1667-1670 DOI:10.1016/j.molp.2016.09.014 |



