2. 冷水性鱼类产业技术创新战略联盟 中国水产科学研究院黑龙江水产研究所 黑龙江 哈尔滨 150070;
3. 安徽省桐城市水产服务中心 安徽 桐城 231400
2. Cold Water Fish Industry Technology Innovation Strategic Alliance, Heilongjiang Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin 150070, China;
3. Tongcheng Fishery Service Center, Tongcheng 231400, China
乌苏里白鲑(Coregonus ussurinsis Berg)属鲑形目(Solmoniformes)、鲑科(Solmonidae)、白鲑亚科(Coregoninae)、白鲑属(Coregonus),主要分布在俄罗斯西伯利亚、萨哈林及我国黑龙江等水域,具有明显的洄游特性,是北方特有冷水性鱼类,也是黑龙江水系中唯一的白鲑属鱼类(张觉民, 1995; 尔斯基, 1960; 罗京等; 2021; 徐革锋等, 2022)。乌苏里白鲑兼具较高的营养和经济价值,是黑龙江名贵水产品之一,但由于生活水体环境恶化、洄游通道受阻及过度捕捞等原因,乌苏里白鲑栖息地逐渐缩减,资源量显著下降,被收录至《中国濒危动物(鱼类)红皮书》中(乐佩琦等, 1998; 徐俐力, 2007; 王继隆等, 2019)。目前,对乌苏里白鲑仅开展了其群体遗传多样性、肌肉营养成分、性腺及胚胎发育、个体繁殖力以及病原体感染等相关研究(董崇智等, 1997; 马波等, 2003; 梁利群等, 2004; 李培伦等, 2015; 李虹娇等, 2017; 王继隆等, 2018; 史秀兰等, 2020; 刘恩慧等, 2022),还缺乏对乌苏里白鲑生长方面的转录组研究,因此,有必要通过RNA-seq测序技术挖掘肌肉生长相关候选基因并阐明其分子调控机制。
鱼类的生长是重要的数量性状,受遗传、环境以及它们间相互作用等多种因素影响,也是养殖鱼类品质评价的重要指标之一(Fuentes et al, 2013; 孙雪等, 2021)。转录组测序(RNA-seq)技术能高效地对遗传调控的相关因子进行深入分析,从分子层面上对调控生长的相关基因进行筛选和鉴定。转录组测序技术已被广泛应用于鱼类生长发育(Al-Tobasei et al, 2017; Paneru et al, 2017; Lu et al, 2020)、免疫应激(Nguyen et al, 2016; Valenzuela-Miranda, 2018; Liu Z et al, 2020; Liang et al, 2021; 朱鑫海等, 2022)和遗传演化(Genet et al, 2014; Kang et al, 2017; Carruthers et al, 2018)等相关基因的挖掘与研究。张波等(2023)从具有生长速度差异的大黄鱼(Larimichthys crocea)群体筛选出3个可能与生长相关的基因;范嗣刚等(2022)从花鲈(Lateolabrax maculatus)中获得10 552个差异表达基因,筛选出igfbp1、fgf、mstn、ghr1等与肌肉生长发育相关的基因;Lu等(2020)在快长和慢长的草鱼(Ctenopharyngodon idella)群体中发现GH/IGF轴、钙代谢、蛋白质和糖原合成、氧转运、细胞骨架和肌原纤维成分通路中的基因参与生长调控;Sun等(2016)发现糖酵解、肌钙蛋白以及参与Ca2+信号传导的基因是杂交石斑鱼(Epinephelus spp.)中的生长优势基因。鱼类生长一直是主要的育种目标性状之一。提高乌苏里白鲑的生长速度不仅可以缩短养殖周期、增加产量,还能够促进其养殖业的发展,提高经济效益的同时满足人们的食用需求,因此,对乌苏里白鲑生长相关研究也变得更加迫切,但目前该鱼的育种工作还停留在传统选育层面,从基因层面对乌苏里白鲑进行选择育种的研究还处于空白。本研究对不同生长速度乌苏里白鲑的肌肉组织进行转录组测序,筛选与生长相关的差异表达基因和信号通路,以期为乌苏里白鲑的分子辅助育种提供基础数据。
1 材料与方法 1.1 实验材料实验用乌苏里白鲑取自中国水产科学研究院黑龙江水产研究所渤海冷水鱼试验站的第2代选育群体,为2018年12月同批次繁育、次年3月破膜成仔鱼、同池混养条件下饲育的3龄鱼。随机选取10尾大规格个体作为快长实验组,体长(24.27±1.48) cm,体重(219.20±38.66) g;随机选取10尾小规格个体作为慢长实验组,体长(17.61±1.80) cm,体重(74.30±17.86) g。将各组实验鱼麻醉后采集背部肌肉组织,置于液氮备用。
1.2 总RNA提取、cDNA文库构建及测序将各实验组中3尾或4尾鱼的背部肌肉等量混合后按照Trizol法提取组织中的总RNA,随后对总RNA进行质量检测:凝胶电泳检测RNA完整性,NanoDrop 2000检验RNA的浓度和纯度,Agilent 2100精确检验RNA的完整性。使用带有Oligo dT的磁珠通过碱基互补的方式富集mRNA,随后将其打断成短片段经PCR扩增得到乌苏里白鲑转录组cDNA文库,共构建6个cDNA文库。将构建合格的cDNA文库使用Illumina高通量测序平台NovaSeq 6000进行测序。转录组文库构建和测序工作委托武汉菲沙基因信息有限公司完成。
1.3 转录组原始数据分析经高通量测序平台获得的原始序列(raw reads)中含有接头信息、低质量及未测出的碱基,不利于转录组后续分析,故而采用SOAPnuke软件(v2.1.0)对原始序列进行质控,即去污染、去接头等处理后获得有效序列(clean reads),随后计算Q20、Q30 (质量值≥20或30的碱基所占百分比)及GC含量。将质控后的有效序列使用HISAT2 (Kim et al, 2015)、bowtie2 (Langmead, 2010)比对软件与本实验室乌苏里白鲑的参考基因组比对(NCBI未发表),进行基因注释。
1.4 样本相关性分析本研究基于基因表达量FPKM (fragments per kilobase per million bases)对样本间的相关性进行分析,以皮尔逊相关系数r2作为判断标准,相关系数越接近1,则样品间相似度越高。
1.5 差异表达基因分析及筛选注释后的基因用FPKM法评估基因表达水平,以慢长组为对照组,快长组为实验组,使用DESeq软件对差异表达基因(DEG)进行筛选,筛选阈值为FDR(false discovery rate) < 0.05且|log2(FC)| > 1(FC, fold change)。基于差异表达基因结果,采用分布(hyper-geometric distribution)的方法对差异表达基因进行GO (gene ontology)功能注释(Ashburner et al, 2000)及KEGG (kyoto encyclopedia of genes and genomes)信号通路富集分析(Kanehisa et al, 2004)。
1.6 qPCR验证对筛选出的10个关键差异表达基因进行qPCR验证。使用反转录试剂盒(TaKaRa, 日本)反转录成cDNA。根据基因CDS序列使用Primer 5.0软件设计qPCR引物,β-actin为内参基因(表 1)。qPCR使用SYBR qPCR Mix试剂盒(EnzyArtisan, 上海)进行操作。PCR扩增体系为10 μL:2 × S6 Universal SYBR qPCR mix 5 μL,正反向引物各0.2 μL,cDNA模板0.5 μL,ddH2O 4.1 μL;扩增程序:95 ℃预变性30 s;42个循环,循环程序为95 ℃变性5 s,60 ℃退火30 s;最后,95 ℃ 15 s,60 ℃ 1 min,95℃ 15 s。使用QuantStudio 6 Flex Real-time PCR仪(Life Technologies, 美国)检测qPCR结果,采用2–ΔΔCt方法计算相对表达量。所有引物委托生工生物工程(上海)股份有限公司合成。
|
|
表 1 qPCR引物序列 Tab.1 qPCR primer sequence |
本研究从构建的6个cDNA文库中共获得295 605 738条原始序列,经质量控制过滤后共获得283 133 612条有效序列,各样品的有效比对率在94.43%以上,Q20的含量在97.80%以上,Q30的含量在93.90%以上,GC碱基含量占总碱基的49.10%以上,无明显GC或AT分离现象。将质控后的有效序列与乌苏里白鲑的参考基因组进行比对,结果显示,比对率在94.93%以上,说明测序结果可靠,可进行后续分析(表 2)。
|
|
表 2 转录组测序质控结果 Tab.2 Quality control results of transcriptome sequencing |
样品相关性系数不仅可以检验生物学实验的可重复性,还可以评估差异表达基因的可靠性,是评估样本选择是否正确的重要指标。本研究通过基因表达量计算各样本之间的相关性系数,并绘制热图(图 1)。由图 1可知,各样本间相关性系数均在0.83以上,表明样本间相关度和实验可靠性较高。
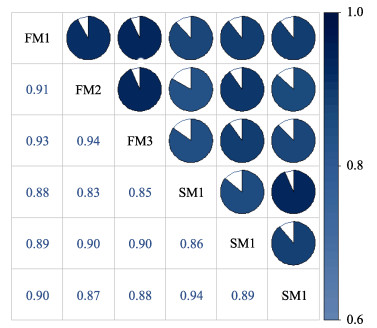
|
图 1 样本相关性分析 Fig.1 Sample correlation analysis |
依据FPKM算法计算基因表达量,使用DESeq软件(v.1.22.2)对基因表达结果进行分析,并计算基因的FC值。以FDR < 0.05且|log2(FC)| > 1作为条件筛选出乌苏里白鲑的生长候选差异基因,并绘制火山图以直观反映快长组和慢长组乌苏里白鲑肌肉组织差异表达基因的分布情况(图 2)。结果显示,在乌苏里白鲑肌肉组织中共鉴定出2 211个差异表达基因,其中包含659个新预测基因;与慢长组相比,快长组有583个差异基因表达上调,1 628个差异基因表达下调。
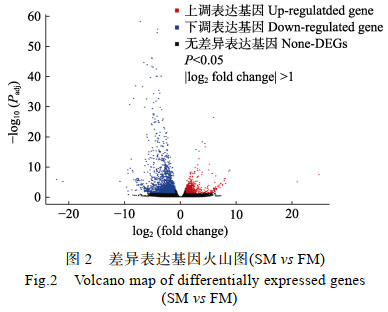
|
图 2 差异表达基因火山图(SM vs FM) Fig.2 Volcano map of differentially expressed genes (SM vs FM) |
GO功能富集分析结果显示,差异表达基因主要富集在生物学过程(biological process, BP)、分子功能(molecular function, MF)和细胞组分(cellular component, CC)的3 620个GO term中,其中,BP最多(2 457个),占67.87%;MF次之(782个),占21.6%;CC最少(381个),占10.52%。由图 3可知,在BP功能中,差异表达基因主要参与细胞过程(cellular process)与代谢过程(metabolic process),分别含有差异基因578和489个;在MF功能中,参与结合(binding)过程的差异表达基因最多,为543个,催化活性(catalytic activity)过程次之,为364个;在CC功能中,差异表达基因主要参与细胞(cell)和细胞组分(cell part)过程,分别包含386和382个差异表达基因。
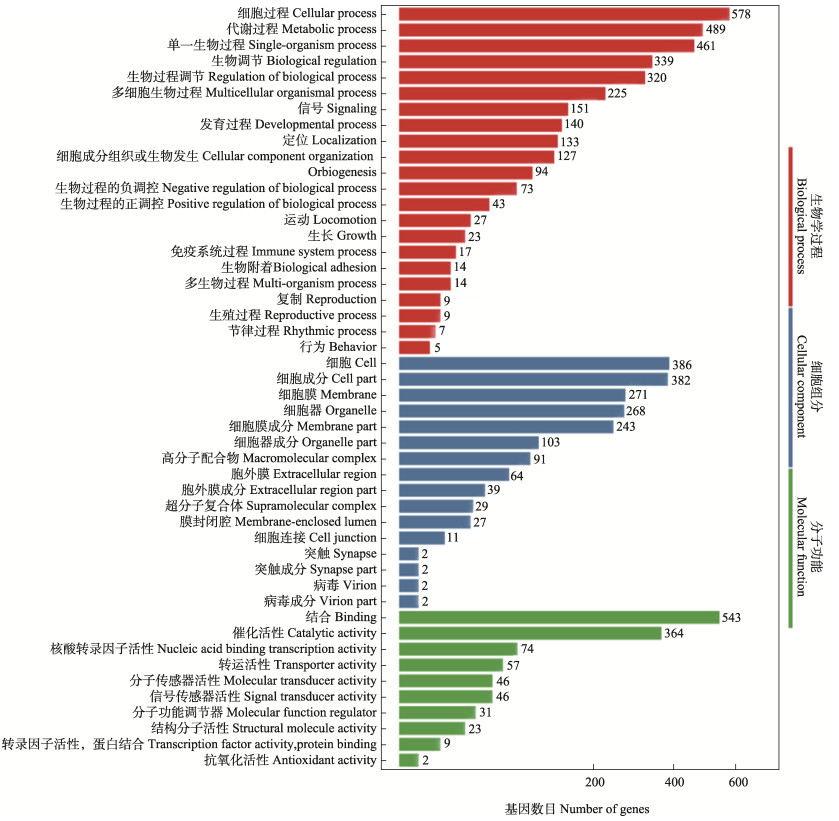
|
图 3 差异表达基因GO功能分类 Fig.3 GO functional classification of differentially expressed genes |
对差异表达基因进行KEGG通路富集分析的结果显示,肌肉组织中的差异表达基因被富集到251条已知KEGG通路中,其中有73条信号通路被显著富集(P < 0.05),快长组中上调的差异表达基因显著富集在糖酵解/糖异生(glycolysis/gluconeogenesis)、甲烷代谢(methane metabolism)、氨基酸的生物合成(biosynthesis of amino acids)、碳代谢(carbon metabolism)以及蛋白酶体(proteasome)通路等(图 4a);下调的差异表达基因显著富集在破骨细胞分化(osteoclast differentiation)、IL-17信号通路(IL-17 signaling pathway)、氨基酸的生物合成(biosynthesis of amino acids)、脂肪细胞因子信号通路(adipocytokine signaling pathway)以及TNF信号通路(TNF signaling pathway)等(图 4b)。
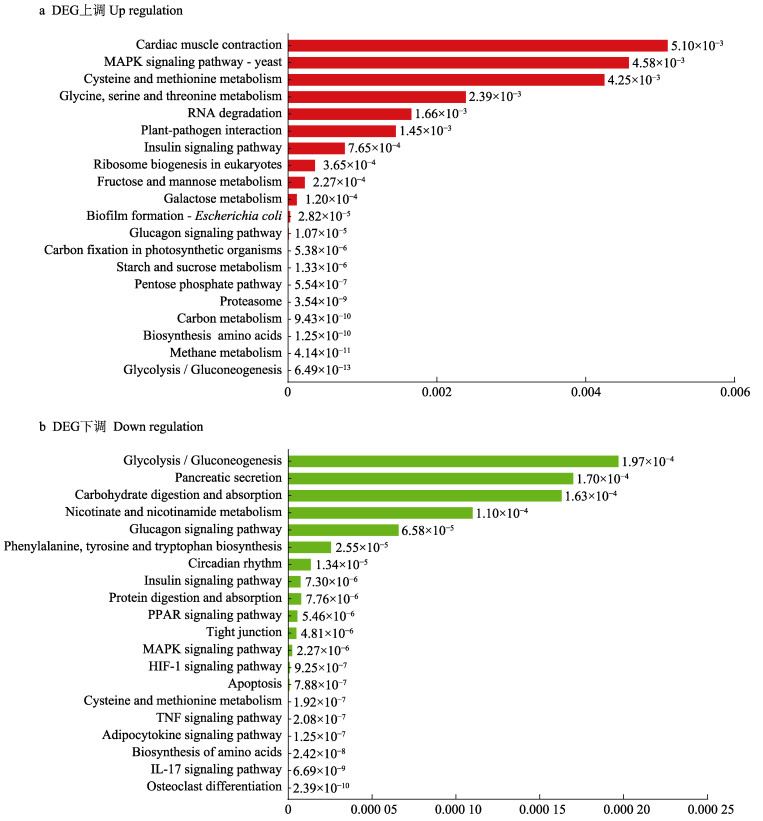
|
图 4 差异表达基因KEGG富集分析 Fig.4 KEGG enrichment analysis of differentially expressed genes |
本研究发现,乌苏里白鲑肌肉生长相关差异基因被显著富集在MAPK信号通路(MAPK signaling pathway)、PI3K-Akt信号通路(PI3K-Akt signaling pathway)、紧密连接(tight junction)、胰岛素信号通路(insulin signaling pathway)、糖酵解/糖异生、PPAR信号通路(PPAR signaling pathway)中(P < 0.05),结合GO功能注释和KEGG通路富集结果,推测差异基因显著富集的信号通路中可能含有与乌苏里白鲑肌肉生长相关的基因,结合相关文献,最终筛选出可能与乌苏里白鲑生长相关的差异基因共31个,如表 3所示。
|
|
表 3 候选差异表达基因注释 Tab.3 Annotation of candidate differentially expressed genes |
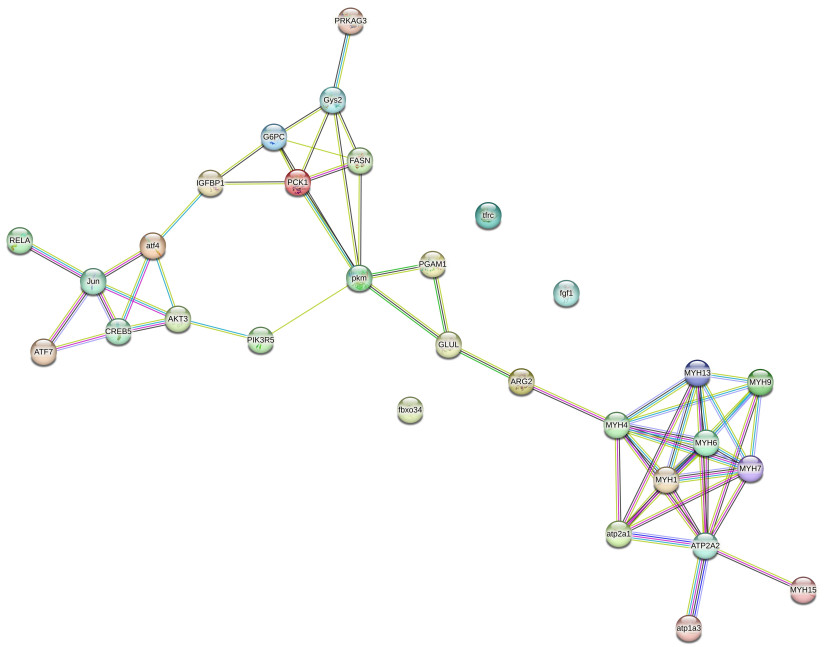
为进一步研究这些差异基因之间的相关作用关系并筛选出可能与乌苏里白鲑肌肉生长相关的关键基因,使用STRING (Version 11.5)数据库(https://string-db.org/)构建蛋白质互作网络图(图 5),除cxcl11.1基因外,在31个差异基因中共鉴定出30个基因编码已知蛋白质,其中27个蛋白质之间存在相互作用。根据蛋白网络互作数筛选出了10个可能与乌苏里白鲑生长相关的关键候选基因:肌浆/内质网钙ATP酶基因atp2a1和atp2a2、葡萄糖-6-磷酸脱氢酶基因g6pc、生长因子结合蛋白1基因igfbp1以及肌球蛋白重链基因myh1、myh4、myh6、myh7、myh9和myh13。其中,快长组中atp2a1、myh1和myh13基因上调表达,atp2a2、g6pci、gfbp1、myh4、myh6、myh7和myh9基因下调表达。
|
图 5 差异基因蛋白互作网络分析 Fig.5 Protein-protein network interaction analysis of differential gene |
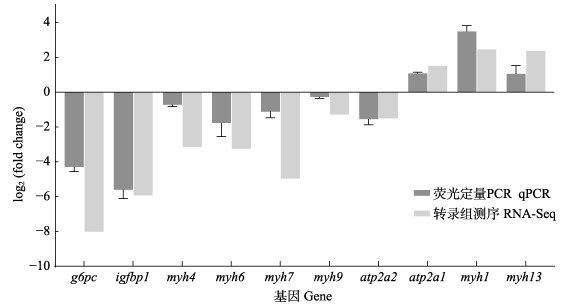
为验证乌苏里白鲑肌肉转录组测序结果的可靠性,对筛选出的10个与生长相关的关键候选基因,以β-actin为内参进行qPCR验证,结果显示(图 6),10个差异基因表达结果与转录组测序获得的基因表达结果趋势一致,证明本研究通过转录组测序获得的结果准确,所筛选出的核心候选基因可靠,可用于后续功能验证和分子标记的开发。
|
图 6 差异表达基因qPCR验证结果 Fig.6 qPCR results of differentially expressed genes |
肌肉生长是评价养殖产品质量和经济效益的重要标准之一,也是优良品种选育过程中不可忽视的指标。乌苏里白鲑是黑龙江省的名贵冷水鱼,因其肉质鲜美广受消费者喜爱。本研究前期发现,同批次繁殖、相同环境条件下养殖的乌苏里白鲑个体规格存在较大差异,为揭示其生长速度显著差异的潜在遗传机制,本研究采用高通量测序技术对快长组和慢长组的乌苏里白鲑进行了转录组测序,筛选出差异表达基因并进行GO和KEGG富集分析,挖掘出了可能与乌苏里白鲑肌肉生长相关的10个候选基因。
atp2a1和atp2a2是ATP2As基因家族的成员,编码负责调节钙跨胞内膜转运的肌浆/内质网Ca2+-ATP酶(SERCA),能够将胞浆中的Ca2+回收至肌浆网腔和内质网腔中,从而肌肉由收缩状态恢复为松弛状态,在控制细胞生长和分化过程中发挥着重要作用(Chemaly et al, 2018)。atp2a1基因在成肌细胞分化过程中表达上调,编码的SERCA1蛋白参与快肌纤维收缩过程,其基因突变显著影响虹鳟(Oncorhynchus mykiss)的生长速度(Salem et al, 2012)。atp2a2基因主要在人体心脏、脑和Ⅰ型骨骼肌(慢肌纤维)发育过程中表达,其编码的SERCA2包含SERCA2a、SERCA2b和SERCA2c三种亚型(Hino et al, 2007; Hovnanian, 2007; Periasamy et al, 2007),atp2a2基因能够减轻小鼠细胞Ca2+失调造成的营养不良(Goonasekera et al, 2011),SERCA含量下降时会延缓心肌舒张,导致心肌收缩功能下降,最终造成心脏衰竭(李红艳等, 2020)。本研究发现,在快长组肌肉组织中atp2a1基因表达量显著上调,atp2a2表达显著下调,可能是通过调节肌浆/内质网中Ca2+通道影响心肌和骨骼肌纤维的收缩与舒张,从而提高乌苏里白鲑神经系统的兴奋性,进而提高乌苏里白鲑的心肌功能与运动能力,通过加速血液循环与骨骼肌收缩对乌苏里白鲑的生长产生了影响。
MYHs基因家族是调控肌肉生长发育的关键基因,其编码的肌球蛋白重链是肌球蛋白(Myosin)Ⅱ类分子中的关键亚基,也是骨骼肌中含量最丰富的蛋白质,在肌肉生长过程中发挥重要作用(陈之航等, 2017);myh在不同类型的骨骼肌纤维中起作用,是肌肉纤维功能特性的主要决定因素,如myh7主要在慢肌纤维中表达,myh1和myh2主要在快肌纤维中表达(Gauvry et al, 1996; Weiss et al, 1999)。本研究发现,在参与乌苏里白鲑生长调节的6个myhs亚型中,快长组中显著上调表达基因为myh1和myh13。myh1及其编码的肌球蛋白-1 (Myosin-1)与脂肪生成有关,其表达上调可增加慢肌纤维的数量,增强肌肉的抗氧化能力,提高运动耐力(Talbot et al, 2016; Ahn et al, 2018; Czapiewski et al, 2022);myh13基因编码特异性眼外肌肌球蛋白重链,参与齿鲸(toothed whales)肌原纤维活性的调节(Senevirathna et al, 2021)。除此之外,本研究中myh4、myh6、myh7和myh9基因在快长组中表达显著下调;myh4基因是成肌分化的标志基因,编码骨骼肌生长所必须的ⅡB型肌球蛋白重链纤维(Lv et al, 2020),myh6基因所编码的肌球蛋白较其他肌球蛋白具有更高的ATP酶活性,在突触传递过程中发挥重要作用(Buga et al, 2008; Miao et al, 2015),myh7基因突变会引起骨骼肌或心肌方面疾病(何一旻等, 2017),同时,推测该基因在肌肉生长发育中具有调控作用(Feinstein-Linial et al, 2016; Zhang et al, 2016);在鱼类中,myh4和myh6基因是参与黑鲷(Acanthopagrus schlegelii)生长调控的关键基因(Lin et al, 2021),myh6和myh7基因在快速生长的兰州鲶(Silurus lanzhouensis)肌肉组织中表达下调(Xiao et al, 2022);myh9基因编码非肌肉肌球蛋白ⅡA重链,且能促进肌纤维分化,其表达量与草鱼(Ctenopharyngodon idellus)肌肉质地有关(Pecci et al, 2018; Xu et al, 2020)。不同亚型的myh基因特异性表达不同肌纤维类型,与快肌纤维相比,慢肌纤维具有更高的有氧运动耐受力和胰岛素敏感性,通过上调快肌表达基因增加快肌纤维的数量及横截面积,提高运动能力,而慢肌纤维能提高肌肉运动的持续时间(Yang et al, 2020)。本研究发现,不同亚型myh基因通过调节其表达影响乌苏里白鲑的肌肉生长,可将MYH基因家族的多个亚型作为乌苏里白鲑生长关键候选基因。
g6pc编码葡萄糖-6-磷酸酶(glucose-6-phosphatase, G-6-pase)催化亚基,该酶是内质网的标志酶,能够催化6-磷酸葡萄糖水解产生葡萄糖和磷酸,是糖酵解和糖异生途径的关键酶,在维持血糖稳态中起着重要作用(钱云霞等, 2011; Liu S et al, 2020)。注射生长抑制素的金钱鱼(Scatophagus argus)体内g6pc基因表达量显著上调,进而激活糖酵解/糖异生过程,消耗大量ATP,导致其生长所需能量不足,从而抑制金钱鱼的生长(Tian et al, 2022)。同样在对草鱼的研究中发现,g6pc基因表达下调能够促进糖原分解从而提高饲料中碳水化合物的利用率(Yue et al, 2021)。g6pc基因主要参与能量与物质代谢过程,在机体代谢和生长的协调过程中发挥重要作用。本研究发现,g6pc基因在快长组的乌苏里白鲑中显著下调,g6pc基因表达能激活糖酵解途径、加速糖原利用、消耗更多能量、减少有机物的积累,该基因高表达时可能会阻碍乌苏里白鲑的生长,可将其作为生长相关候选基因,进一步对其进行功能验证及分子标记的挖掘。
IGF结合蛋白(IGFBPs)和IGF、IGF受体(IGFR)以及IGFBP水解酶四部分共同构成胰岛素样生长因子(insulin-like growth factors, IGFs),IGFs参与细胞分裂与分化、生物体代谢、生长与繁殖等过程,是GH/IGF生长轴(生长激素/胰岛素样生长因子轴)的关键因子,也是动物生长发育过程中的重要调控因子(黄建峰等, 2011; Hakuno et al, 2015)。IGFBPs是一种分泌性蛋白,在人和哺乳动物中已鉴定出6种亚型(IGFBP1~IGFBP6),它与IGF形成复合体后可延长IGF的半衰期,并激活胰岛素受体,抑制低血糖的发生(Duan et al, 2005; 赵艳等, 2015)。igfbp1是IGFBPs家族中第一个被发现和鉴定的成员,其结构与功能已在多个物种中进行克隆和研究,其表达水平与鲑鱼的体重、体长、生长速率等指标呈负相关(Shimizu et al, 2006);在对虹鳟(Kocmarek et al, 2014)和斑马鱼(Danio rerio) (Opazo et al, 2017)的研究中发现,igfbp1基因均在小规格个体中表达上调。本研究发现,与慢长组相比,快长组igfbp1基因表达下调,这与斑马鱼、虹鳟等研究结果一致,可将其作为乌苏里白鲑生长相关候选基因进行后续验证。
4 结论本研究通过对不同生长速度的乌苏里白鲑肌肉组织进行转录组测序,共发现了2 211个差异表达基因,其中包含659个新预测基因;583个差异基因在快长组中表达显著上调,1 628个差异基因表达显著下调。此外,GO和KEGG富集分析发现,与生长相关的基因被显著富集在MAPK信号通路、PI3K-Akt信号通路、紧密连接、胰岛素信号通路、糖酵解/糖异生、PPAR信号通路中。进一步对筛选出候选差异表达基因进行PPI蛋白互作网络分析,确定了10个与生长相关的核心候选基因:atp2a2、atp2a1、g6pc、igfbp1、myh1、myh4、myh6、myh7、myh9和myh13。本研究为乌苏里白鲑生长性状相关的分子调控机制及分子标记辅助育种的研究提供了基础资料。
AHN J S, KIM D H, PARK H B, et al. Ectopic overexpression of porcine Myh1 increased in slow muscle fibers and enhanced endurance exercise in transgenic mice. International Journal of Molecular Sciences, 2018, 19(10): 2959 DOI:10.3390/ijms19102959 |
AL-TOBASEI R, ALI A, LEEDS T D, et al. Identification of SNPs associated with muscle yield and quality traits using allelic-imbalance analyses of pooled RNA-Seq samples in rainbow trout. BMC Genomics, 2017, 18(1): 582 DOI:10.1186/s12864-017-3992-z |
ASHBURNER M, BALL C A, BLAKE J A, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics, 2000, 25(1): 25-29 DOI:10.1038/75556 |
BUGA A M, SASCAU M, PISOSCHI C, et al. The genomic response of the ipsilateral and contralateral cortex to stroke in aged rats. Journal of Cellular and Molecular Medicine, 2008, 12(6b): 2731-2753 DOI:10.1111/j.1582-4934.2008.00252.x |
CARRUTHERS M, YURCHENKO A A, AUGLEY J J, et al. De novo transcriptome assembly, annotation and comparison of four ecological and evolutionary model salmonid fish species. BMC Genomics, 2018, 19(1): 32 DOI:10.1186/s12864-017-4379-x |
CHEMALY E R, TRONCONE L, LEBECHE D. SERCA control of cell death and survival. Cell Calcium, 2018, 69: 46-61 DOI:10.1016/j.ceca.2017.07.001 |
CHEN Z H, DONG J J, SUN C F, et al. cDNA cloning and analyses of two myosin heavy chain isoforms of mandarin fish (Siniperca chuatsi) based on transcriptome sequencing. Progress in Fishery Sciences, 2017, 38(3): 51-61 [陈之航, 董浚键, 孙成飞, 等. 基于转录组测序对翘嘴鳜(Siniperca chuatsi) 2种肌球蛋白重链基因的克隆与分析. 渔业科学进展, 2017, 38(3): 51-61] |
CZAPIEWSKI R, BATRAKOU D G, DE LAS HERAS J I, et al. Genomic loci mispositioning in Tmem120a knockout mice yields latent lipodystrophy. Nature Communications, 2022, 13(1): 321 DOI:10.1038/s41467-021-27869-2 |
DONG C Z, XIA Z Z, JIANG Z F, et al. The tentative studies on reproduction population structure of Coregonus ussurinsis. Chinese Journal of Fisheries, 1997, 10(1): 14-21 [董崇智, 夏重志, 姜作发, 等. 黑龙江乌苏里白鲑生殖群体生态学特征及资源保护. 水产学杂志, 1997, 10(1): 14-21] |
DUAN C, XU Q. Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. General and Comparative Endocrinology, 2005, 142(1/2): 44-52 |
FAN S G, YANG W Y, HUANG H, et al. Muscle transcriptome analysis and growth-related genes screening of Lateolabrax maculatus with different growth rate. Journal of Guangdong Ocean University, 2022, 42(5): 9-17 [范嗣刚, 杨文燕, 黄皓, 等. 不同生长速率花鲈肌肉转录组分析及生长相关基因筛选. 广东海洋大学学报, 2022, 42(5): 9-17 DOI:10.3969/j.issn.1673-9159.2022.05.002] |
FEINSTEIN-LINIAL M, BUVOLI M, BUVOLI A, et al. Two novel MYH7 proline substitutions cause Laing Distal Myopathy-like phenotypes with variable expressivity and neck extensor contracture. BMC Medical Genetics, 2016, 17(1): 57 DOI:10.1186/s12881-016-0315-1 |
FUENTES E N, VALDÉS J A, MOLINA A, et al. Regulation of skeletal muscle growth in fish by the growth hormone--insulin-like growth factor system. General and Comparative Endocrinology, 2013, 192: 136-148 DOI:10.1016/j.ygcen.2013.06.009 |
GAUVRY L, FAUCONNEAU B. Cloning of a trout fast skeletal myosin heavy chain expressed both in embryo and adult muscles and in myotubes neoformed in vitro. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 1996, 115(2): 183-190 DOI:10.1016/0305-0491(96)00074-0 |
GENET C, VERRIER E R, CIOBOTARU C, et al. RNA-Seq analysis of transcriptome response to VHS-V infection in two target tissues of resistant vs susceptible trout clonal lines. Proceedings of the 10th World Congress on Genetics Applied to Livestock Production, Vancouver, BC, Canada, 2014, 17–22
|
GOONASEKERA S A, LAM C K, MILLAY D P, et al. Mitigation of muscular dystrophy in mice by SERCA overexpression in skeletal muscle. Journal of Clinical Investigation, 2011, 121(3): 1044-1052 DOI:10.1172/JCI43844 |
HAKUNO F, FUKUSHIMA T, YONEYAMA Y, et al. The novel functions of high-molecular-mass complexes containing insulin receptor substrates in mediation and modulation of insulin-like activities: Emerging concept of diverse functions by IRS-associated proteins. Frontiers in Endocrinology, 2015, 6: 73 |
HE Y M, GU M M. Research progress of myosin heavy chain genes in human genetic diseases. Hereditas (Beijing), 2017, 39(10): 877-887 [何一旻, 顾鸣敏. 肌球蛋白重链基因在人类遗传性疾病中的研究进展. 遗传, 2017, 39(10): 877-887] |
HINO S I, KONDO S, SEKIYA H, et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Human Molecular Genetics, 2007, 16(23): 2834-2843 DOI:10.1093/hmg/ddm239 |
HOVNANIAN A. SERCA pumps and human diseases. Sub-Cellular Biochemistry, 2007, 45: 337-363 |
HUANG J F, LIAO Z Y. Structure and physiological function of IGFs. Science and Technology Association Forum, 2011(3): 59-60 [黄建峰, 廖志勇. 胰岛素样生长因子结构与生理功能. 科协论坛(下半月), 2011(3): 59-60 DOI:10.3969/j.issn.1007-3973.2011.03.036] |
KANEHISA M, GOTO S, KAWASHIMA S, et al. The KEGG resource for deciphering the genome. Nucleic Acids Research, 2004, 32(1): D277-D280 |
KANG J, MA X, HE S. Evidence of high-altitude adaptation in the glyptosternoid fish, Creteuchiloglanis macropterus from the Nujiang River obtained through transcriptome analysis. BMC Evolutionary Biology, 2017, 17(1): 1-12 DOI:10.1186/s12862-016-0855-1 |
KIM D, LANGMEAD B, SALZBERG S L. HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 2015, 12(4): 357-360 DOI:10.1038/nmeth.3317 |
KOCMAREK A L, FERGUSON M M, DANZMANN R G. Differential gene expression in small and large rainbow trout derived from two seasonal spawning groups. BMC Genomics, 2014, 15(1): 57 DOI:10.1186/1471-2164-15-57 |
LANGMEAD B. Aligning short sequencing reads with Bowtie. Current Protocols Bioinformatics, 2010, Chapter 11: Unit 11.7
|
LI H J, HAN Y. Investigation report on biological characteristics Coregonus ussurinsis. Northern Chinese Fisheries, 2017(1): 17-20 [李虹娇, 韩英. 乌苏里白鲑生物学特征调查报告. 黑龙江水产, 2017(1): 17-20 DOI:10.3969/j.issn.1674-2419.2017.01.007] |
LI H Y, ZHAO S H, WANG S H, et al. Research progress of SERCA2a in heart failure. Chinese Pharmacological Bulletin, 2020, 36(2): 171-174 [李红艳, 赵思涵, 王世华, 等. SERCA2a在心力衰竭中的研究进展. 中国药理学通报, 2020, 36(2): 171-174 DOI:10.3969/j.issn.1001-1978.2020.02.006] |
LI P L, LIU W, WANG J L, et al. Fecundity of Coregonus ussurinsis in the Heilongjiang River, China. Journal of Fishery Sciences of China, 2015, 22(6): 1234-1242 [李培伦, 刘伟, 王继隆, 等. 黑龙江乌苏里白鲑的个体繁殖力. 中国水产科学, 2015, 22(6): 1234-1242] |
LIANG L Q, CHANG Y M, DONG C Z. Analysis of genetic diversity for Coregonus ussurinsis Berg in Heilongjiang River. Journal of Fishery Sciences of China, 2004, 11(6): 501-505 [梁利群, 常玉梅, 董崇智. 黑龙江乌苏里白鲑遗传多样性分析. 中国水产科学, 2004, 11(6): 501-505 DOI:10.3321/j.issn:1005-8737.2004.06.003] |
LIANG P, SAQIB H S A, LIN Z, et al. RNA-seq analyses of marine medaka (Oryzias melastigma) reveals salinity responsive transcriptomes in the gills and livers. Aquatic Toxicology, 2021, 240: 105970 DOI:10.1016/j.aquatox.2021.105970 |
LIN Z, ZHANG Z, SOLBERG M F, et al. Comparative transcriptome analysis of mixed tissues of black porgy (Acanthopagrus schlegelii) with differing growth rates. Aquaculture Research, 2021, 52(11): 5800-5813 DOI:10.1111/are.15455 |
LIU E H, HUANG T Q, GU W, et al. Cloning and tissue expression of liver-expressed antimicrobial peptide Leap-2 in Ussuri cisco Coregonus ussurinsis exposed to bacterial infection. Journal of Dalian Ocean University, 2022, 37(3): 420-427 [刘恩慧, 黄天晴, 谷伟, 等. 乌苏里白鲑肝脏表达抗菌肽Leap-2基因的克隆及细菌感染后的组织表达分析. 大连海洋大学学报, 2022, 37(3): 420-427] |
LIU S, TIAN F, ZHANG C, et al. Genome-wide identification, phylogeny and expression analysis of G6PC gene family in common carp, Cyprinus carpio. Turkish Journal of Biochemistry, 2020, 45(2): 205-212 DOI:10.1515/tjb-2018-0102 |
LIU Z, ZHAO L, HUANG L, et al. Integration of RNA-seq and RNAi provides a novel insight into the immune responses of Epinephelus coioides to the impB gene of Pseudomonas plecoglossicida. Fish and Shellfish Immunology, 2020, 105: 135-143 DOI:10.1016/j.fsi.2020.06.023 |
LU X, CHEN H M, QIAN X Q, et al. Transcriptome analysis of grass carp (Ctenopharyngodon idella) between fast-and slow-growing fish. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 2020, 35: 100688 DOI:10.1016/j.cbd.2020.100688 |
LUO J, HAN Y. Research progress of Second Songhua River fishery resources. Northern Chinese Fisheries, 2021, 40(2): 3-7 [罗京, 韩英. 第二松花江渔业资源研究进展. 黑龙江水产, 2021, 40(2): 3-7 DOI:10.3969/j.issn.1674-2419.2021.02.001] |
LV W, JIN J, XU Z, et al. lncMGPF is a novel positive regulator of muscle growth and regeneration. Journal of Cachexia, Sarcopenia and Muscle, 2020, 11(6): 1723-1746 DOI:10.1002/jcsm.12623 |
MA B, SHI L Y, DONG C Z. Biochenical genetic structure in Coregonus ussurinsis Berg. Journal of Fishery Sciences of China, 2003, 10(3): 195-200 [马波, 石连玉, 董崇智. 乌苏里白鲑的生化遗传结构. 中国水产科学, 2003, 10(3): 195-200 DOI:10.3321/j.issn:1005-8737.2003.03.004] |
MIAO Y, YANG J, XU Z, et al. RNA sequencing identifies upregulated kyphoscoliosis peptidase and phosphatidic acid signaling pathways in muscle hypertrophy generated by transgenic expression of myostatin propeptide. International Journal of Molecular Sciences, 2015, 16(4): 7976-7994 DOI:10.3390/ijms16047976 |
NGUYEN T V, JUNG H, NGUYEN T M, et al. Evaluation of potential candidate genes involved in salinity tolerance in striped catfish (Pangasianodon hypophthalmus) using an RNA-Seq approach. Marine Genomics, 2016, 25: 75-88 DOI:10.1016/j.margen.2015.11.010 |
NIKOLSKY B. Translated by GAO X. Fishes of Amur River. Beijing: Science Press, 1960 [尼科尔斯基. 译者: 高岫. 黑龙江流域鱼类. 北京: 科学出版社, 1960]
|
OPAZO R, VALLADARES L, ROMERO J. Comparison of gene expression patterns of key growth genes between different rate growths in zebrafish (Danio rerio) siblings. Latin American Journal of Aquatic Research, 2017, 45(4): 766-775 DOI:10.3856/vol45-issue4-fulltext-12 |
PANERU B D, AL-TOBASEI R, KENNEY B, et al. RNA-Seq reveals MicroRNA expression signature and genetic polymorphism associated with growth and muscle quality traits in rainbow trout. Scientific Reports, 2017, 7(1): 1-15 DOI:10.1038/s41598-016-0028-x |
PECCI A, MA X, SAVOIA A, et al. MYH9: Structure, functions and role of non-muscle myosin ⅡA in human disease. Gene, 2018, 664: 152-167 DOI:10.1016/j.gene.2018.04.048 |
PERIASAMY M, KALYANASUNDARAM A. SERCA pump isoforms: Their role in calcium transport and disease. Muscle Nerve, 2007, 35(4): 430-442 DOI:10.1002/mus.20745 |
QIAN Y X, ZHENG W X, SONG J J. Cloning and sequence analysis of Lateolabrax japonicus glucose-6-phosphatase catalytic subunit (G6PC) cDNA and its 5'-flanking region. Journal of Agricultural Biotechnology, 2011, 19(4): 606-615 [钱云霞, 郑伟贤, 宋娟娟. 鲈鱼6-磷酸葡萄糖酶催化亚基(G6PC) cDNA和5'侧翼序列的克隆及分析. 农业生物技术学报, 2011, 19(4): 606-615 DOI:10.3969/j.issn.1674-7968.2011.04.003] |
SALEM M, VALLEJO R L, LEEDS T D, et al. RNA-Seq identifies SNP markers for growth traits in rainbow trout. PLoS One, 2012, 7(5): e36264 DOI:10.1371/journal.pone.0036264 |
SENEVIRATHNA J D, YONEZAWA R, SAKA T, et al. Transcriptomic insight into the melon morphology of toothed whales for aquatic molecular developments. Sustainability, 2021, 13(24): 13997 DOI:10.3390/su132413997 |
SHI X L, WNAG B Q, HAUNG T Q, et al. Observation on embryo development of Ussuri whitefish Coregonus ussuriensis Berg in Heilongjiang River. Oceanologia et Limnologia Sinica, 2020, 51(2): 415-421 [史秀兰, 王炳谦, 黄天晴, 等. 黑龙江乌苏里白鲑(Coregonus ussuriensis Berg)胚胎发育观察研究. 海洋与湖沼, 2020, 51(2): 415-421] |
SHIMIZU M, BECKMAN B R, HARA A, et al. Measurement of circulating salmon IGF binding protein-1: Assay development, response to feeding ration and temperature, and relation to growth parameters. Journal of Endocrinology, 2006, 188(1): 101-110 DOI:10.1677/joe.1.06475 |
SUN X, LI S J, DU J X, et al. The effects of pyramiding growth-related genotypes in grass carp (Ctenopharyngodon idella). Progress in Fishery Sciences, 2021, 42(5): 40-46 [孙雪, 李胜杰, 杜金星, 等. 草鱼生长相关优势基因型的聚合效果分析. 渔业科学进展, 2021, 42(5): 40-46] |
SUN Y, HUANG Y, HU G, et al. Comparative transcriptomic study of muscle provides new insights into the growth superiority of a novel grouper hybrid. PLoS One, 2016, 11(12): e0168802 DOI:10.1371/journal.pone.0168802 |
TALBOT J, MAVES L. Skeletal muscle fiber type: Using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdisciplinary Reviews: Developmental Biology, 2016, 5(4): 518-534 DOI:10.1002/wdev.230 |
TIAN C, ZHANG J, FENG P, et al. Comparative analysis of transcriptome responses to injected somatostatin 3 peptide in spotted scat (Scatophagus argus). Aquaculture Reports, 2022, 23: 101022 DOI:10.1016/j.aqrep.2022.101022 |
VALENZUELA-MIRANDA D, GALLARDO-ESCÁRATE C. Dual RNA-Seq uncovers metabolic amino acids dependency of the intracellular bacterium Piscirickettsia salmonis infecting Atlantic salmon. Frontiers in Microbiology, 2018, 9: 2877 DOI:10.3389/fmicb.2018.02877 |
WANG J L, LIU W, LI P L, et al. Evaluation of nutritive quality and nutrient components in the muscle of Coregonus ussuriensis.. Journal of Guangdong Ocean University, 2018, 38(5): 35-40 [王继隆, 刘伟, 李培伦, 等. 乌苏里白鲑肌肉营养成分与品质评价. 广东海洋大学学报, 2018, 38(5): 35-40] |
WANG J L, LIU W, LU W Q, et al. Assessment of the population resources of Coregonus ussuriensis in the middle reaches of Amur River. Chinese Journal of Ecology, 2019, 38(6): 1824-1829 [王继隆, 刘伟, 鲁万桥, 等. 黑龙江中游乌苏里白鲑资源现状评估. 生态学杂志, 2019, 38(6): 1824-1829] |
WEISS A, MCDONOUGH D, WERTMAN B, et al. Organization of human and mouse skeletal myosin heavy chain gene clusters is highly conserved. Proceedings of the National Academy of Sciences, 1999, 96(6): 2958-2963 DOI:10.1073/pnas.96.6.2958 |
XIAO W, LIAN Z Q, WU J P, et al. De novo RNA sequencing for identification of growth-related genes in Silurus lanzhouensis muscle tissues. Fisheries Science, 2022, 88(5): 565-580 DOI:10.1007/s12562-022-01598-x |
XU G F, WANG B Q, LI M S. The fish that knows how to go home for the winter - Coregonus ussuriensis. China Fisheries, 2022(3): 112-114 [徐革锋, 王炳谦, 李明爽. 懂得回家"猫冬"的鱼——乌苏里白鲑. 中国水产, 2022(3): 112-114] |
XU L L. Survival status of several rare cold-water fishes in Heilongjiang Province. Scientific and Technological Innovation, 2007(13): 137 [徐俐力. 黑龙江省几种珍稀冷水鱼类生存现状. 黑龙江科技信息, 2007(13): 137] |
XU W H, GUO H H, CHEN S J, et al. Transcriptome analysis revealed changes of multiple genes involved in muscle hardness in grass carp (Ctenopharyngodon idellus) fed with faba bean meal. Food Chemistry, 2020, 314: 126205 DOI:10.1016/j.foodchem.2020.126205 |
YANG H, LI X Q, XU Z, et al. Effects of three active components in Eucommia ulmoides on growth and flesh quality of grass carp (Ctenopharyngodon idellus) based on transcriptomics. Aquaculture Nutrition, 2020, 26(6): 1895-1907 DOI:10.1111/anu.13109 |
YUE D, HUANG S, YANG R, et al. Effects of tea polyphenols on the growth performance, carbohydrate metabolism of grass carp (Ctenopharyngodon idellus). Aquaculture Nutrition, 2021, 27(6): 2344-2354 DOI:10.1111/anu.13367 |
YUE P Q, CHEN Y Y. China red book of endangered animals (fish). Beijing: Science Press, 1998 [乐佩琦, 陈宜瑜. 中国濒危动物(鱼类)红皮书. 北京: 科学出版社, 1998]
|
ZHANG B, JIANG D, ZHANG D L, et al. Comparative analysis of transcriptome of muscle tissue of individuals with different growth rate of Larimichthys crocea. Journal of Fisheries of China, 2023, 47(3): 87-100 [张波, 姜丹, 张东玲, 等. 大黄鱼生长速率差异个体肌肉组织的转录组的比较分析. 水产学报, 2023, 47(3): 87-100] |
ZHANG J M. Fishes of Heilongjiang Province. Heilongjiang: Heilongjiang Science and Technology Press, 1995 [张觉民. 黑龙江省鱼类志. 黑龙江: 黑龙江科学技术出版社, 1995]
|
ZHANG Z, JIANG X, LI Q, et al. Differential expression of MYH7 gene in different tissues of chicken. China Poultry, 2016, 38(24): 52-54 |
ZHAO Y, LU L, LIU Y Z, et al. Research progress on physiological function and expression regulation of insulin-like growth factor-binding protein-1. Shandong Agricultural Sciences, 2015, 47(1): 139-143 [赵艳, 卢玲, 刘云章, 等. Igfbp-1的生理功能及其表达调控的研究进展. 山东农业科学, 2015, 47(1): 139-143] |
ZHU X H, ZHANG Z R, ZHOU L Y, et al. Transcriptomic analysis of the head kidney of Siniperca chuatsi infected with Aeromonas hydrophila. Progress in Fishery Sciences, 2022, 43(4): 208-217 [朱鑫海, 张紫瑞, 周丽颖, 等. 嗜水气单胞菌感染翘嘴鳜头肾转录组分析. 渔业科学进展, 2022, 43(4): 208-217] |



