2. 海南热带海洋学院崖州湾创新研究院 海南 三亚 572025;
3. 中国海洋大学 海水养殖教育部重点实验室 山东 青岛 266003;
4. 海南省热带海水养殖工程技术研究中心 海南 海口 571126
2. Yazhou Bay Innovation Institute, Hainan Tropical Ocean University, Sanya 572025, China;
3. Ocean University of China, Key Laboratory of Mariculture, Ministry of Education, Qingdao 266003, China;
4. Hainan Provincial Engineering Research Center for Tropical Sea-Farming, Haikou 571126, China
石斑鱼广泛分布于全球热带、亚热带的暖水性中下层水域,是一种珊瑚礁鱼类。石斑鱼营养丰富、市场价值高、消费需求大,为大宗海洋经济鱼类,深受消费者欢迎,养殖潜力巨大(Ge et al, 2019)。《中国渔业统计年鉴》数据显示,我国石斑鱼养殖产量逐年上升,2021年产量超过20万t,同比上一年增长6.29%(农业农村部渔业渔政管理局等, 2022),位居我国海水养殖鱼类产量前十,我国现已成为石斑鱼主产国(Rimmer et al, 2019)。然而,随着养殖规模的扩大,石斑鱼产业出现了种质退化、优质苗种匮乏等诸多问题(Shapawi et al, 2019),亟需开展其重要经济性状遗传解析和良种选育工作。目前,石斑鱼的良种选育主要采用选择育种和杂交育种等传统方式(Rimmer et al, 2019)。随着分子生物技术的快速发展和高通量测序成本的下降,SSR(micro-satellite)、SNP (single nucleotide polymorphism)等分子标记相继出现(黎裕等, 1999),分子标记开发逐渐成为石斑鱼性状解析和遗传育种的重要手段。其中,微卫星因具有遗传共显性、多态性高、信息含量丰富、种间可转移性好、可视化和基因组覆盖率高等优点(Deng et al, 2014),被广泛应用于物种区分、群体遗传结构分析、遗传多样性评估、亲子鉴定和标记辅助选择育种等研究。
石斑鱼作为重要的海水经济鱼类,其分子标记被广泛开发并应用。樊欣等(2022)利用SSR标记,鉴定了斜带石斑鱼(Epinephelus coioides)人工雌核生殖子代的遗传物质来源。Xiao等(2018)结合荧光AFLP标记技术揭示了七带石斑鱼(Hyporthodus septemfascia-tus)养殖群体的遗传多样性和种群遗传结构。蒙子宁等(2007)利用RAPD技术分析了斜带石斑鱼繁育亲本与子代的遗传多样性。唐江等(2018)利用微卫星标记分析了新品种云龙石斑鱼[鞍带石斑鱼(E.lanceolatus) (♂)×云纹石斑鱼(E.moara)(♀)]与其父母本群体间的遗传变异情况。目前,石斑鱼分子标记开发数量有限,在育种上的应用也鲜有报道。因此,开发大量具有实用价值的分子标记对提高石斑鱼育种效率具有重大意义。
近年来,随着高通量测序技术的快速发展和测序成本的下降,全基因组和转录组分析促进了石斑鱼分子标记的开发(Tóth et al, 2000; 高峰涛等, 2017)。目前,赤点石斑鱼(E.akaara)、斜带石斑鱼、棕点石斑鱼(E. fuscoguttatus)、鞍带石斑鱼、云纹石斑鱼已完成全基因组测序。为开发与经济性状相关联的分子标记,构建分子辅助育种技术体系,本研究基于全基因组数据对5种石斑鱼SSR进行筛选,并对其种类、数量进一步比较分析,以期为具有生长、抗逆和抗病等性状的石斑鱼新品种选育奠定基础。
1 材料与方法 1.1 微卫星位点筛选从CNGB (https://www.cngb.org)、NCBI (https://www.ncbi.nlm.nih.gov/)、DDBJ (https://www.ddbj.nig.ac.jp)和Dryad (https://datadryad.org/stash)数据库中获取5种石斑鱼的基因组序列(Ge et al, 2019; Zhou et al, 2019、2021; Yang et al, 2021),详细信息见表 1。利用Micro-Satellite (MISA)软件(https://webblast.ipk-gatersleben.de/misa/) (Beier et al, 2017)进行5种石斑鱼基因组微卫星序列的筛选:重复单元:1~6 bp,单碱基重复拷贝数≥12,二碱基重复拷贝数≥6,三、四、五和六碱基重复拷贝数均≥5。
|
|
表 1 5种石斑鱼全基因组信息 Tab.1 The information of whole genome in the five species of grouper |
导出MISA检索数据,利用自编脚本整合数据,挑选完美型微卫星,利用Excel软件统计5种石斑鱼基因组完美型微卫星序列总长度、相对丰度(个/Mb;微卫星数量/基因组总长度)、SSR平均距离(bp/个;基因组总长度/微卫星数量)、重复单元类型[微卫星重复拷贝序列中的碱基,如(AT)16中的AT]、重复类型(单、二、三、四、五和六碱基重复)、核心拷贝数[重复单元出现的次数,如:(AT)16中的16]、重复拷贝类别(表示重复类别具体由哪些碱基组成)。根据碱基互补配对原则和起始碱基顺序的排列差异,将同类重复兼并为同一种重复拷贝类别,如AGC类别包括AGC、GCA、CAG、GCT、TGC和CTG,AGCG类别包括GCGA、CGAG、GAGC、AGCG、TCGC、CTCG、GCTC和CGCT (Jurka et al, 1995)。
2 结果与分析 2.1 5种石斑鱼基因组各重复类型微卫星总体分布特征从公共数据库中获取5种石斑鱼全基因组数据(表 1),使用MISA软件与自编脚本筛选整合其完美型微卫星序列,统计结果见表 2。5种石斑鱼基因组大小均超过1G,微卫星相对丰度介于271~296个/Mb之间,总长度为6.30~7.06 Mb,平均长度为22 bp左右,各统计参数总体差别不大,分布规律一致。5种石斑鱼各重复类型的数量、占比、相对丰度排序一致,皆为二碱基重复最多,其次为单碱基,并随着重复单元碱基数目的增加而减少。
|
|
表 2 5种石斑鱼微卫星数量及分布特征 Tab.2 Amount and distribution of microsatellite in the five species of grouper genome |
通过分析5种石斑鱼基因组中各重复拷贝类别的特征发现,不同和相同重复类别的数量、占比稍有差异,但整体趋势一致(表 3和表 4)。单碱基重复类别中,A、C类别间数目差距巨大,A类别重复的数量在5种石斑鱼基因组中占据绝对优势,在该类别中占比达90.00%左右(表 3)。在二碱基重复类别中,AC类别数量占据绝对优势,在该重复类别中占比接近80.00%,同时,是所有重复拷贝类别中数量最多的类别(表 4);CG类别含量最少,在5种石斑鱼二碱基重复类别中仅占0.04%~0.10%。三碱基重复类别中,数量占前三的类别为AAT、AGG和AGC,比例分别为29.62%~31.41%、17.38%~18.69%、12.55%~16.26%。四碱基重复类别中,AATC、AAAT、AGAT和AATG为优势类别,4种类别数量排序稍有不同,但差距不大;其中,云纹石斑鱼中AATC类别的含量位居所有重复类别数量前十名。五碱基优势重复类别为AGAGG、AAAAT和AAGAT;其中,赤点石斑鱼的重复类别数量排序与其他4种鱼类相比有些许差别。六碱基优势重复类别各不相同,但多为AAANNN (N代表除A以外的其他3种碱基)、AANNNN和ACAGAG。
|
|
表 3 5种石斑鱼各重复类型中前3种优势重复拷贝类别 Tab.3 The top three dominant duplicated copy categories for each duplication type in the five species of grouper genome |
|
|
表 4 5种石斑鱼中出现次数最多的10种重复拷贝类别的计数和百分比 Tab.4 The counts and percentage of most frequent top 10 duplicated copy categories observed in the five species of grouper |
通过分析5种石斑鱼基因组微卫星重复拷贝数,发现不同重复类型微卫星的拷贝数变化具有显著差异,但每种重复类型的重复拷贝数变化趋势一致,微卫星数量均随拷贝数的增加而减少(图 1)。5种石斑鱼(赤点石斑鱼、斜带石斑鱼、棕点石斑鱼、鞍带石斑鱼和云纹石斑鱼)单碱基重复拷贝数均集中在12~25次(图 1a),分别占单碱基重复数量的99.35%、96.54%、97.54%、95.39%和97.34%。二碱基重复中,拷贝数主要集中在6~32次之间(图 1b),占二碱基重复数量的97.53%、98.91%、98.58%、99.59%和97.79%;其中,分布数量在11~14次之间小幅度上涨。在三碱基重复中,拷贝数主要集中于5~16次(图 1c),分别占99.33%、99.18%、99.65%、98.99%和99.34%。四碱基重复拷贝数主要集中在5~17次之间(图 1d),分别占99.51%、99.41%、99.36%、99.81%和99.65%;其中,AGAT和AAAG重复拷贝数较大时分布数量较多。五碱基重复中,拷贝数主要集中在5~14次(图 1e),分别占98.21%、99.06%、99.52%、99.90%和99.56%;其中,AAGAG、AATAT和AGAGG重复拷贝数较大时,分布数量较多。在六碱基重复中,拷贝数主要集中于5~12次之间(图 1f),分别占100%、99.60%、100%、99.68%和99.75%。
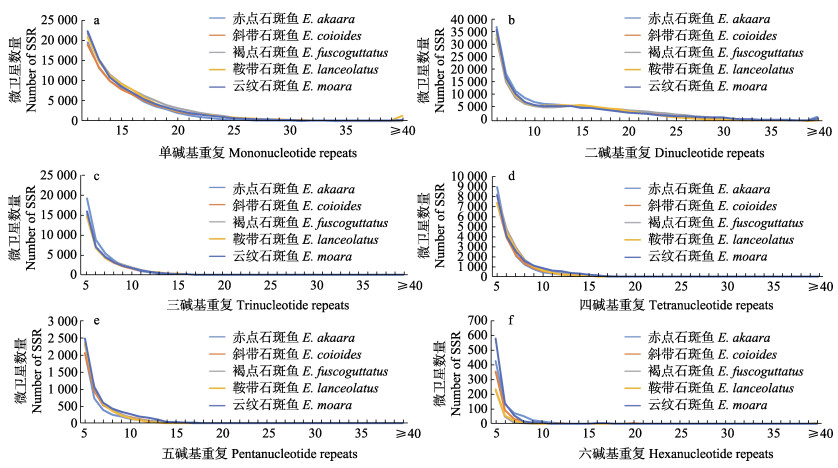
|
图 1 5种石斑鱼全基因组中微卫星不同重复类型拷贝数分布 Fig.1 Distribution of copy numbers in different microsatellites repeat types in the whole genomes of five species of groupers |
不同重复类型微卫星的重复拷贝数分布显示(图 2),拷贝数在6和12次时微卫星分布数量出现峰值,并随着拷贝数的增加而递减;其中,微卫星数量在重复拷贝数为6次时最多,其数量在4~5万之间,分别占总微卫星数量的16.75%、15.77%、14.78%、15.80%和16.00%。此外,重复拷贝数最大的SSR位点均分布于鞍带石斑鱼和云纹石斑鱼中。鞍带石斑鱼中重复拷贝数最大的位点为单碱基重复T、二碱基重复TA和六碱基重复AGACAG,分别重复502、803和48次。云纹石斑鱼的三碱基重复GAG、四碱基重复CACT、五碱基重复CCACA为重复拷贝数最大的位点,分别重复205、652和111次。
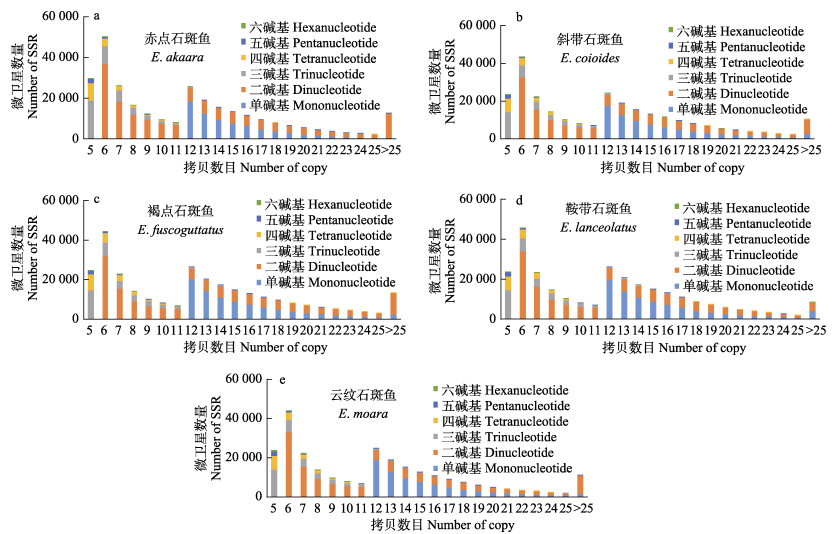
|
图 2 5种石斑鱼微卫星各重复拷贝数分布特征 Fig.2 The distribution characteristics of copy numbers of microsatellite repeat in five species of groupers |
随着下一代测序(next generation sequencing, NGS)技术的进步,全基因组、转录组序列等数据迅速积累,越来越多的微卫星被快速识别并应用。军曹鱼(Rachycentron canadum) (马骞等, 2023)、团头鲂(Megalobrama amblycephala) (张芹等, 2022)、凡纳对虾(Penaeus vannamei) (王佳佳等, 2023)、红鳍东方鲀(Takifugu rubripes) (崔建洲等, 2006)、卵形鲳鲹(Trachinotus ovatus) (张永德等, 2020)、黄颡鱼(Pelteobagrus fulvidraco)(徐杰杰等, 2021)、金钱鱼(Scatophagus argus) (王耀嵘等, 2020)、斑鳢(Channa maculata) (上官清等, 2020)等都进行了基因组微卫星分子标记的筛选。本研究利用已发布的石斑鱼全基因组数据,筛选出大量微卫星位点。相比于RFLP (restriction fragment length polymorphism) (Ramirez et al, 2006)和AFLP (amplified fragment length polymorphism) (Xiao et al, 2018)等一次性仅能开发9个和600个微卫星标记的传统方式,全基因组数据筛选微卫星数量多达28万个以上,数量更多,经济快捷,且覆盖范围广。
3.1 5种石斑鱼微卫星全基因组分布分析5种石斑鱼全基因组数据筛选出的微卫星序列总长分别占各自基因组序列的0.59%~0.67% (表 2),低于人类(Homo sapiens)(3%)(Subramanian et al, 2003)、牛(Bos taurus)(4.7%)、绵羊(Ovis aries)(4.8%) (戚文华等, 2013)等哺乳动物所占比例;但棕点石斑与大熊猫(Ailuropoda melanoleuca)(0.64%)(李午佼等, 2014)相比,所占比例较高。这种现象可能归因于基因组大小和基因组内碱基组成及排列方式的差异(Hancock, 1996)。在鱼类中,黄颡鱼(1.8%)(徐杰杰等, 2020)、斑点叉尾鮰(Ictalurus punctatus)(1.45%)(唐荣叶等, 2022)、草鱼(Ctenopharyngodon idella)(1.43%)(黄纬杰等, 2022)、红鳍东方鲀(0.77%)(崔建洲等, 2006)等鱼类微卫星在基因组序列中的比例均比石斑鱼高,表明5种石斑鱼基因组重复序列中可能存在稀疏性(Xu et al, 2020)。
在5种石斑鱼基因组中,除单碱基外,随着微卫星重复单元碱基数目的增加,微卫星分布频率均逐渐下降。与斑点叉尾鮰(唐荣叶等, 2022)、斑鳢(上官清等, 2020)、草鱼(黄纬杰等, 2022)等鱼类及与牛、绵羊(戚文华等, 2013)、大熊猫、北极熊(Ursus maritimus) (李午佼等, 2014)等哺乳动物中单碱基重复的微卫星优势类型不同,5种石斑鱼基因组中二碱基微卫星数目最多,占微卫星总数的49.02%~54.00%。导致此现象的原因之一是所用微卫星筛选软件及筛选规则不同。本研究采用碱基数目均≥12 bp原则,而其他动物的筛选规则采用单碱基从10 bp开始筛选(若本研究采用≥10 bp筛选原则,则优势重复类型为单碱基)。但在豚鹿(Axis porcinus)(耿广耀等, 2022)等哺乳动物和金钱鱼(王耀嵘等, 2020)、凡纳对虾(王佳佳等, 2023)等水生动物中,二碱基微卫星是数量最具优势的碱基类型,与本研究结果一致。由此可见,不同物种的优势微卫星重复类型不同,而物种在微卫星分布上的差异一定程度上可以表现为物种基因组的进化现象。虽然优势类型不同,但各重复类型分布比例未有较大变化,表现出微卫星的进化保守性。
3.2 各重复拷贝类别微卫星特征分析5种石斑鱼中,A类别的数量在单碱基重复中占据绝对优势,其比例为90%左右,与人类、大熊猫、北极熊、黑青斑河豚(Tetraodon nigroviridis)、斑点叉尾鮰、草鱼等情况相似。这可能与上述动物中大量穿插的Alu和LINE-1(Long interspersed nuclear elements-1)等反转录序列和假基因有关(Edwards et al, 1998)。在二碱基重复中,AC、AG、AT类别在5种石斑鱼该重复拷贝类别的微卫星分布数量排行前三,而CG类别的含量最低,仅占据二碱基重复数量的0.04%~ 0.10%,CG类别重复在大部分物种中数量都极少(Tóth et al, 2000)。这是因为不同物种基因组内4种碱基组成含量本身具有差异性,另一方面也可能是不同碱基存在结构上的问题(Edwards et al, 1998)。在三碱基重复中,排名前三的类别均为AAT、AGG和AGC,其中,AAT的高频率分布与人类、大熊猫、凡纳滨对虾、草鱼、红鳍东方鲀等大多数动物一致,AGG与AGC在大多数物种中分布数量较少。有研究表明,AGG是参与其他物种早期生长发育的许多转录因子已知结合位点,而AGG类别中的GGA和GAA重复序列能减弱大鼠(Rattus norvegicus)多聚免疫球蛋白受体基因的表达调控(徐杰杰等, 2021)。除此以外,AGC类别的碱基重复多态性变化与遗传疾病直接相关,具有进化和医学研究意义。研究发现,在AGC类别的CTG重复序列中,CTG作为肌营养不良症肌强直蛋白激酶(DMPK)基因3´UTR区的组成成分,其异常扩增会引起人Ⅰ型强直性肌营养不良症(DM1)(Brook et al, 1992),同时出现男性不育、白内障、智力障碍、心律不齐等症状(Ergoli et al, 2020)。因此,AGG和AGC类别的高频率分布很可能在石斑鱼免疫、疾病等基因的表达调控中发挥重要作用。在四、五、六碱基重复类别中,AAAN、AAAAN和AAAAAN类别均为各碱基重复类别中优势碱基类别的前3名,其中,AAAN类别在哺乳动物中含量十分丰富(Tóth et al, 2000);而AAAAN在灵长类动物中分布较多,在啮齿类动物中较少。
3.3 不同微卫星重复类型的拷贝数特征分析5种石斑鱼全基因组中,各重复类型的微卫星总数均在重复拷贝数为6或12次时出现峰值,并均随着重复拷贝数的增加,微卫星分布数逐渐减少(图 2)。这与绒杜父鱼(Hemitripterus villosus)、金钱鱼、卵形鲳鲹等鱼类全基因组中微卫星的变化规律一致。这种各重复类型微卫星含量随序列长度变化的规律性现象,主要是由于微卫星的序列长度与其稳定性呈负相关,而微卫星的突变率与其重复拷贝数呈正相关,即微卫星序列越长,其稳定性降低,突变频率升高,导致数量越少(Wierdl et al, 1997);另一方面,单碱基和二碱基分别从≥12和≥6次重复开始筛选,因此,在这2个点形成了峰值。由此可见,不同的筛选原则导致不同物种得到的统计结果不同。然而,目前没有统一的微卫星筛选原则,在各个物种比较时出现结果与实际不符或无法比较的情况。
在比较5种石斑鱼各重复类型中重复拷贝数的分布时发现,鞍带石斑鱼(T、TA和AGACAG)和云纹石斑鱼(GAG、CACT、和CCACA)存在重复拷贝数最大的微卫星位点。有研究表明,AT是高等真核生物中最具扩展性的重复类型,反映了鞍带石斑鱼和云纹石斑鱼对应位点上的微卫星在进化过程中可能更容易扩张(Tóth et al, 2000)。该重复位点也可能在物种进化和生长等自身发展起到重要作用,对物种的适应性和生存能力产生影响。其中,GAG重复序列是胃癌中具有高度微卫星不稳定性的突变靶点(Ge et al, 2019),其多态性的变化可能会导致疾病的发生。重复拷贝数的增加可能代表着相应功能的变化,因此,对核心拷贝数最高的重复类型进行进一步的研究,有助于探索石斑鱼的进化机制和功能表达。
目前,少见关于石斑鱼全基因组筛选微卫星的报道,而利用其他方式筛选出的微卫星标记多应用于石斑鱼遗传多样性的研究(Xiao et al, 2018; 蒙子宁等, 2007),在育种方面的应用研究较少。本研究利用生物信息学软件,在5种石斑鱼全基因组中筛选符合条件的微卫星,为进一步研究其基因组特征、进化、表达奠定基础。同时,利用在石斑鱼全基因组中筛选得到的微卫星位置信息,可为批量开发微卫星引物,为石斑鱼功能标记筛选、亲子鉴定及新品种培育等育种工作提供基础资料。
BEIER S, THIEL T, MÜNCH T, et al. MISA-web: A web server for microsatellite prediction. Bioinformatics, 2017, 33(16): 2583-2585 DOI:10.1093/bioinformatics/btx198 |
BROOK J D, MCCURRACH M E, HARLEY H G, et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3'end of a transcript encoding a protein kinase family member. Cell, 1992, 68(4): 799-808 DOI:10.1016/0092-8674(92)90154-5 |
Bureau of Fisheries, Ministry of Agriculture and Rural Affairs, National Fisheries Technology Extension Center, China Society of Fisheries. China fishery statistical yearbook 2022. Beijing: China Agriculture Press, 2022 [农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会. 2022 中国渔业统计年鉴. 北京: 中国农业出版社, 2022]
|
CUI J Z, SHEN X Y, YANG G P, et al. The analysis of simple sequence repeats in Takifugu rubripes genome. Periodical of Ocean University of China (Natural Science), 2006, 36(2): 249-254 [崔建洲, 申雪艳, 杨官品, 等. 红鳍东方鲀基因组微卫星特征分析. 中国海洋大学学报(自然科学版), 2006, 36(2): 249-254] |
DENG Y W, LEI Q N, TIAN Q L, et al. De novo assembly, gene annotation, and simple sequence repeat marker development using Illumina paired-end transcriptome sequences in the pearl oyster Pinctada maxima. Bioscience, Biotechnology, and Biochemistry, 2014, 78(10): 1685-1692 DOI:10.1080/09168451.2014.936351 |
EDWARDS Y J, ELGAR G, CLARK M S, et al. The identification and characterization of microsatellites in the compact genome of the Japanese puffer fish, Fugu rubripes: Perspectives in functional and comparative genomic analyses. Journal of Molecular Biology, 1998, 278(4): 843-854 DOI:10.1006/jmbi.1998.1752 |
ERGOLI M, VENDITTI M, PICILLO E, et al. Study of expression of genes potentially responsible for reduced fitness in patients with myotonic dystrophy type 1 and identification of new biomarkers of testicular function. Molecular Reproduction and Development, 2020, 87(1): 45-52 DOI:10.1002/mrd.23307 |
FAN X, ZHANG W W, WU Y C, et al. Introduction of diploid gynogenesis in orange-spot grouper Epinephelus coioidesl by giant grouper Epinephelus lanceolatus sper. Acta Hydrobiologica Sinica, 2022, 46(11): 1675-1683 [樊欣, 张维炜, 吴廷昌, 等. 鞍带石斑鱼精子诱导斜带石斑鱼雌核生殖. 水生生物学报, 2022, 46(11): 1675-1683 DOI:10.7541/2022.2021.0241] |
GAO F T, SHAO C W, CUI Z K, et al. Development and population genetic diversity analysis of microsatellite markers in Epinephelus awoara. Periodocal of Ocean University of China (Natural Science), 2017, 47(4): 52-57 [高峰涛, 邵长伟, 崔忠凯, 等. 基于高通量测序的青石斑鱼基因组微卫星开发及评价. 中国海洋大学学报(自然科学版), 2017, 47(4): 52-57] |
GE H, LIN K B, SHEN M, et al. De novo assembly of a chromosome-level reference genome of red-spotted grouper (Epinephelus akaara) using nanopore sequencing and Hi-C. Molecular Ecology Resources, 2019, 19(6): 1461-1469 DOI:10.1111/1755-0998.13064 |
GENG G Y, YOU Y Y, LIU Q X. SSR molecular marker development and genetic diversity analysis of Axis porcinus. Chinese Journal of Wildlife, 2022, 43(3): 816-820 [耿广耀, 由玉岩, 刘群秀. 豚鹿SSR标记开发与遗传多样性分析. 野生动物学报, 2022, 43(3): 816-820 DOI:10.12375/ysdwxb.20220329] |
HANCOCK J M. Simple sequences in a "minimal" genome. Nature Reviews Genetics, 1996, 14(1): 14-15 DOI:10.1038/ng0996-14 |
HUANG W J, GUO X Z, ZHANG Z H, et al. Analysis of microsatellite in the entire grass carp (Ctenopharyngodon idella) genome and the application in parentage identification. Journal of Fisheries of China, 2022, 46(2): 161-172 [黄纬杰, 郭向召, 张子豪, 等. 草鱼全基因组微卫星特征分析与亲子鉴定. 水产学报, 2022, 46(2): 161-172] |
JURKA J, PETHIYAGODA C. Simple repetitive DNA sequences from primates: Compilation and analysis. Journal of Molecular Evolution, 1995, 40(2): 120-126 DOI:10.1007/BF00167107 |
LI W J, LI Y Z, DU L M, et al. Comparative analysis of microsatellite sequences distribution in the genome of giant panda and polar bear. Sichuan Journal of Zoology, 2014, 33(6): 874-878 [李午佼, 李玉芝, 杜联明, 等. 大熊猫和北极熊基因组微卫星分布特征比较分析. 四川动物, 2014, 33(6): 874-878] |
LI Y, JIA J Z, WANG T Y. Types of molecular markers and their development. Biotechnology Information, 1999, 15(4): 21-24 [黎裕, 贾继增, 王天宇. 分子标记的种类及其发展. 生物技术通报, 1999, 15(4): 21-24] |
MA Q, WU Y W, WANG L Y, et al. Screening and characterization of polymorphic SSR markers based on whole genome sequencing of cobia (Rachycentron canadum). Progress in Fishery Sciences, 2023, 44(4): 135-144 [马骞, 吴雨薇, 王刘永, 等. 军曹鱼全基因组微卫星特征分析与多态性标记的筛选及应用. 渔业科学进展, 2023, 44(4): 135-144] |
MENG Z N, YANG L P, WU F, et al. Genetic diversity in cultured stocks of Epinephelus coioides by RAPD analysis. Journal of Tropical Oceanography, 2007, 26(2): 44-48 [蒙子宁, 杨丽萍, 吴丰, 等. 斜带石斑鱼养殖群体遗传多样性的RAPD分析. 热带海洋学报, 2007, 26(2): 44-48 DOI:10.3969/j.issn.1009-5470.2007.02.008] |
QI W H, JIANG X M, XIAO G S, et al. Seeking and bioinformatics analysis of microsatellite sequence in the genomes of cow and sheep. Acta Veterinaria et Zootechnica Sinica, 2013, 44(11): 1724-1733 [戚文华, 蒋雪梅, 肖国生, 等. 牛和绵羊全基因组微卫星序列的搜索及其生物信息学分析. 畜牧兽医学报, 2013, 44(11): 1724-1733] |
RAMIREZ M A, PATRICIA-ACEVEDO J, PLANAS S. New microsatellite resources for groupers (Serranidae). Molecular Ecology Notes, 2006, 6(3): 813-817 DOI:10.1111/j.1471-8286.2006.01354.x |
RIMMER M A, GLAMUZINA B. A review of grouper (Family Serranidae: Subfamily Epinephelinae) aquaculture from a sustainability science perspective. Reviews in Aquaculture, 2019, 11: 58-87 DOI:10.1111/raq.12226 |
SHANGGUAN Q, CHEN K C, LIU H Y, et al. Characteristics of micorsatellites and genetic structure of wild Channa maculate. South China Fisheries Science, 2020, 16(3): 47-60 [上官清, 陈昆慈, 刘海洋, 等. 斑鳢基因组中微卫星分布特征及野生种群遗传结构分析. 南方水产科学, 2020, 16(3): 47-60] |
SHAPAWI R, ABDULLAH F C, SENOO S, et al. Nutrition, growth and resilience of tiger grouper (Epinephelus fuscoguttatus)×giant grouper (Epinephelus lanceolatus) hybrid: A review. Reviews in Aquaculture, 2019, 11(4): 1285-1296 DOI:10.1111/raq.12292 |
SUBRAMANIAN S, MISHRA R K, SINGH L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biology, 2003, 4(2): R13 DOI:10.1186/gb-2003-4-2-r13 |
TANG J, TIAN Y S, LI Z T, et al. Analysis of genetic characters in Epinephelus moara, E. lanceolaus and their hybrids. Journal of Agricultural Biotechnology, 2018, 26(5): 819-829 [唐江, 田永胜, 李振通, 等. 云纹石斑鱼和鞍带石斑鱼及其杂交后代遗传性状分析. 农业生物技术学报, 2018, 26(5): 819-829] |
TANG R Y, SU M Y, YANG W S, et al. Analysis of microsatellite distribution characteristics in the channel catfish (Ictalurus punctatus) genome. Progress in Fishery Sciences, 2022, 43(2): 89-97 [唐荣叶, 苏孟园, 杨汶珊, 等. 斑点叉尾鮰全基因组微卫星分布特征分析. 渔业科学进展, 2022, 43(2): 89-97] |
TÓTH G, GÁSPÁRI Z, JURKA J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Research, 2000, 10(7): 967-981 DOI:10.1101/gr.10.7.967 |
WANG J J, WANG Q, QIN Z, et al. Development of SSR markers from genomic data for Litopenaeus vannamei and analysis of genetic diversity in different cultured populations. Journal of Fisheries of China, 2023, 47(6): 64-74 [王佳佳, 王琼, 秦桢, 等. 凡纳滨对虾全基因组SSR标记开发及不同养殖群体的遗传多样性. 水产学报, 2023, 47(6): 64-74] |
WANG Y R, YANG W, REN X L, et al. Distribution patterns of microsatellites and development of polymorphic markers from Scatophagus argus genome. Journal of Guangdong Ocean University, 2020, 40(4): 7-14 [王耀嵘, 杨尉, 任席林, 等. 金钱鱼基因组微卫星分布特征分析及多态性标记开发. 广东海洋大学学报, 2020, 40(4): 7-14] |
WIERDL M, DOMINSKA M, PETES T D. Microsatellite instability in yeast: Dependence on the length of the microsatellite. Genetics, 1997, 146(3): 769-779 DOI:10.1093/genetics/146.3.769 |
XIAO Y S, GUAN S G, LIU Q H, et al. Genetic characteristics of broodstock and offspring of the seven-band grouper (Hyporthodus septemfasciatus ) using fluorescent-AFLP markers. Journal of the Marine Biological Association of the United Kingdom, 2018, 98(2): 261-267 DOI:10.1017/S0025315416001260 |
XU J J, ZHENG X, LI J, et al. Distribution characteristics of whole genome microsatellite of Pelteobagrus fulvidraco. Genomics and Applied Biology, 2020, 39(12): 5488-5498 [徐杰杰, 郑翔, 李杰, 等. 黄颡鱼(Pelteobagrus fulvidraco)全基因组微卫星分布特征分析. 基因组学与应用生物学, 2020, 39(12): 5488-5498] |
XU J J, ZHENG X, ZHANG X Y, et al. Analysis of distribution characteristics of microsatellites in four genomes of puffer fish. Genomics and Applied Biology, 2021, 40(4): 1441-1451 [徐杰杰, 郑翔, 张鑫宇, 等. 4种河鲀全基因组微卫星分布特征分析. 基因组学与应用生物学, 2021, 40(4): 1441-1451] |
YANG Y, WANG T, CHEN J F, et al. Whole-genome sequencing of brown-marbled grouper (Epinephelus fuscoguttatus) provides insights into adaptive evolution and growth differences. Molecular Ecology Resources, 2021, 22(2): 711-723 |
ZHANG Q, WANG Y H, WANG B K, et al. Characterization of genomic microsatellites and development of SSR markers of Megalobrama amblycephala. Jiangsu Agricultural Sciences, 2022, 50(3): 79-85 [张芹, 王延晖, 王冰柯, 等. 团头鲂基因组微卫星特征及SSR位点开发. 江苏农业科学, 2022, 50(3): 79-85] |
ZHANG Y D, WEN L T, LUO H L, et al. Genome survey and development of SSR molecular markers for Trachinotus ovatus. Journal of Southern Agriculture, 2020, 51(5): 983-994 [张永德, 文露婷, 罗洪林, 等. 卵形鲳鲹基因组调研及其SSR分子标记的开发应用. 南方农业学报, 2020, 51(5): 983-994] |
ZHOU Q, GAO H Y, XU H, et al. A chromosomal-scale reference genome of the kelp grouper Epinephelus moara. Marine Biotechnology, 2021, 23(1): 12-16 |
ZHOU Q, GAO H Y, ZHANG Y, et al. A chromosome-level genome assembly of the giant grouper (Epinephelus lanceolatus) provides insights into its innate immunity and rapid growth. Molecular Ecology Resources, 2019, 19(5): 1322-1332 |



