2. 烟台经海海洋渔业有限公司 山东 烟台 264006;
3. 安徽省桐城市水产服务中心 安徽 桐城 231400
2. Yantai Jinghai Marine Fishery Co. LTD, Yantai 264006, China;
3. Anhui Tongcheng Aquatic Products Service Center, Tongcheng 231400, China
虹鳟(Oncorhynchus mykiss)是目前世界上养殖范围最广的冷水性经济鱼类之一。我国自20世纪60年代起,陆续从朝鲜、美国、丹麦等国家引进虹鳟种质,在种质保存、鉴定、养殖和品种选育等方面开展了系统研究,目前,已选育出生长速度快、适合我国养殖模式的虹鳟“水科1号”新品种(王炳谦等, 2021)。随后虹鳟“全雌1号”通过了全国水产原种和良种审定委员会审定,实现了核心种源自主可控。三倍体虹鳟饲料转化率高、生长优势明显,肉质优于二倍体,备受国内外养殖户和消费者的欢迎。同时,三倍体性腺发育不全(Han et al, 2010; Huang et al, 2021; Krisfalusi et al, 1999; Nynca et al, 2022),避免了其产卵期死亡率高和肉质品质下降的养殖风险(Cleveland et al, 2016; Manor et al, 2012、2014; Werner et al, 2008),并且规避了养殖逃逸造成外来物种入侵等生态风险(Glover et al, 2016; Karlsson et al, 2016; Nie et al, 2014; Xu et al, 2022),因此,养殖三倍体虹鳟具有更为显著的市场效益和生态效益。
由于虹鳟二倍体和三倍体在形态学上非常相似,很难通过形态学来区分虹鳟倍性。以往多倍体通过细胞学分析直接计数染色体数目来确定,但这种方法往往会导致虹鳟死亡(苏泽古等, 1984; 尹洪滨, 2001; 印杰等, 2005)。近年来,多采用流式细胞术测定细胞中DNA的相对含量来确定样本的倍性。然而,使用流式细胞术对样本血细胞分析来检测倍性,采集血液的鱼类体长至少需5 cm,并且这可能会对鱼体造成伤害(Allen, 1983; Lecommandeur et al, 1994; 齐嫣然等, 2022)。此外,这两种方法操作精细、技术流程复杂、耗材昂贵,因此,虹鳟三倍体倍性鉴定的普及应用受到了极大制约。
微卫星标记(SSR标记)是广泛分布在真核生物基因组中的简单重复序列,已广泛应用于鱼类的倍性和谱系分析(Ohara et al, 1999、2000; Mishina et al, 2014; Nie et al, 2014)。微卫星分析的优势包括重现性高、样本需求量低及群体遗传分析快速等。微卫星分析已应用于亚马逊帆鳍鳉(Poecilia formosa) (Lampert et al, 2006)、银鲫(Carassius auratus)(Bai et al, 2011)、大菱鲆(Scophthalmus maximus) (Hernández-Urcera et al, 2012)、大西洋鲑(Salmo salar) (Glover et al, 2016)和泥鳅(Misgurnus anguillicaudatus) (Feng et al, 2018)的多倍体鉴定。因此,本研究采用微卫星技术分析虹鳟二倍体群体和三倍体群体间的差异,以期为养殖条件下区分二倍体和三倍体虹鳟提供理论依据。
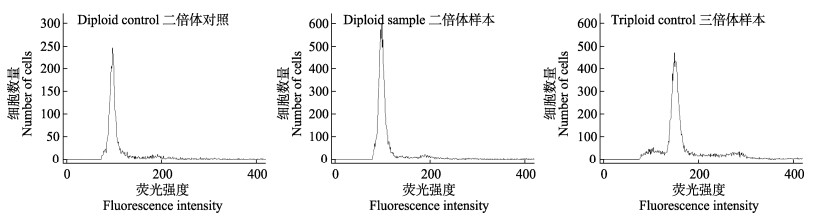
1 材料与方法 1.1 样本采集实验鱼来自中国水产科学研究院黑龙江水产研究所渤海冷水性鱼试验站,鱼体长(15±2) cm,体重(30±5) g。共采集52尾已知倍性(26尾二倍体,26尾三倍体)和48尾未知倍性(二倍体和三倍体混合比例未知)虹鳟幼鱼血液及鳍条组织作为样本。其中52尾虹鳟幼鱼的倍性通过使用流式细胞仪分析红细胞DNA含量来确定(图 1)。
|
图 1 二倍体和三倍体样本(细胞DNA含量)的流式细胞仪检测结果(以2n=30的虹鳟红细胞作为二倍体对照) Fig.1 Diploid and triploid samples (cell DNA content) were detected by flow cytometry, red blood cells of rainbow trout (2n=30) as diploid control |
使用DNA提取试剂盒(Tiangen)从虹鳟尾鳍中提取DNA。在紫外分光度计上测定纯度(OD260/280=1.8~2.0)和浓度,并将浓度稀释到50 ng/µL,使用1%琼脂糖凝胶电泳检测质量。
1.3 PCR扩增及检测153个虹鳟微卫星标记由Rexroad、Coulibaly和Phillips等开发(Rexroad et al, 2001、2002a、b、c; Coulibaly et al, 2005; Phillips et al, 2006),利用primer3对其中部分SSR标记重新设计了稳定的引物(Robinson et al, 2004),由生工生物(上海)工程公司合成。
PCR总体系为10 µL,包括2×Tap Mix 5 µL;上、下游引物(10 µmol/L)各0.5 µL;DNA模板1 µL;ddH2O 3 µL。在ABI 9700型PCR仪进行如下PCR:94 ℃预变性5 min;94 ℃变性30 s,退火温度58 ℃复性30 s,72 ℃延伸30 s,25个循环;最后72 ℃延伸7 min。扩增产物经8%聚丙烯酰胺凝胶电泳分离、银染法显色,数码相机拍摄保存。
1.4 筛选出的SSR标记在倍性鉴定中的有效性验证使用选择的SSR标记对来自中国水产科学研究院黑龙江水产研究所渤海冷水性鱼试验站的48个未知倍性的虹鳟样本进行验证。每个个体采集尾鳍提取DNA;尾静脉穿刺法采集血液进行流式细胞术分析。利用筛选所得的SSR标记进行PCR扩增。根据每个个体在SSR位点上的最大等位基因数来估计倍性。
为了验证倍性检测的准确性,采用sysmex流式细胞仪测量每个个体的DNA含量,并以2n=30的虹鳟红细胞作为二倍体对照。
1.5 数据统计与分析使用Gel-Pro Analyzer (Version 6.3)软件进行图像数据分析,获得特异等位基因及基因型数据,并统计等位基因片段大小,分析和计算每个标记在二倍体、三倍体群体的等位基因数(A)、观测杂合度(Ho)。观测杂合度计算公式:观测杂合度(Ho)=观察得到杂合个体数/样本个体数总数。
2 结果与分析 2.1 微卫星标记对虹鳟基因组DNA的扩增结果及遗传差异本研究使用的153个SSR标记,139个(90.85%) SSR标记在二倍体虹鳟基因组DNA中扩增得到清晰条带;141个(92.16%) SSR标记在三倍体虹鳟基因组DNA中扩增得到清晰条带;139个(90.85%) SSR标记在二倍体和三倍体虹鳟基因组DNA中均扩增得到清晰条带,其中,132个(86.27%)标记在个体间表现出多态。
132个具有多态性的SSR标记的样本PCR扩增片段在79~482 bp,每个标记检测到的等位基因数在1~18之间,共检测到588个等位基因,二倍体群体410个,三倍体群体402个。二倍体群体在139个标记的等位基因数(A)在1~14之间,平均为3.11,三倍体群体的A值介于1~16之间,平均为3.05。二倍体群体观测杂合度(Ho)在0.00~1.00之间,平均为0.66;三倍体群体的Ho在0.00~1.00之间,平均为0.71。两个群体遗传多样性处于较高水平。
2.2 虹鳟倍性鉴定特征性位点筛选首先,通过使用流式细胞仪分析红细胞DNA含量来确定52尾虹鳟幼鱼的倍性,区分出26尾二倍体样本和26尾三倍体样本(图 1)。使用153个SSR标记对4个(2个二倍体和2个三倍体)DNA样本进行PCR扩增,产物经聚丙烯酰胺凝胶分离后,检测到132个标记具有多态性。通过观察等位基因数目来计算等位基因实际数目。对表现出多态的132个标记进行倍性鉴定特征性位点的第一次筛选。倍性鉴定特征性位点的筛选标准为:所有二倍体样本的图谱条带数目≤2,三倍体样本中包含个体图谱条带数目≥3。经过图像数据分析,13个SSR标记符合筛选标准。
随后将进行PCR的DNA样本数量增加到48个(24个二倍体和24个三倍体),对13个符合初次筛选标准的SSR标记进行第二次筛选。结果显示,7个SSR标记(表 1)符合,其余6个SSR标记不符合筛选标准的主要原因是个别二倍体样本的图谱条带数目>2。
|
|
表 1 符合虹鳟倍性鉴定特征性位点筛选标准的SSR引物及遗传参数 Tab.1 SSR primers and genetic parameters conforming to the screening criteria of characteristic sites for ploidy identification of rainbow trout |
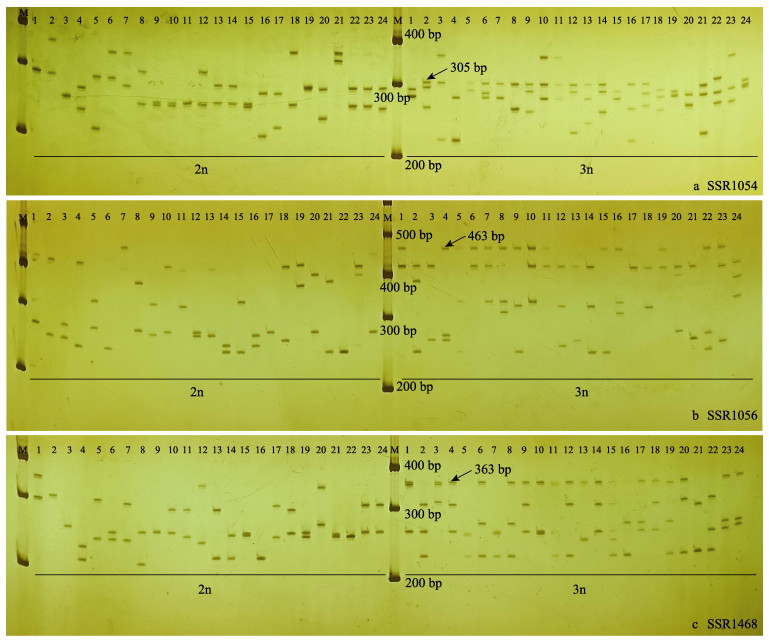
从符合倍性鉴定特征性位点筛选标准的7个SSR标记中选择3个SSR引物(SSR1054、SSR1056和SSR1468)作为鉴别倍性水平的标记(表 2),它们不仅具有较多的等位基因数和较高的观测杂合度,而且在这3个标记均检测出三倍体特有的等位基因(表 3),如图 2所示,SSR1054-305bp、SSR1056-463bp和SSR 1 468-363bp 3条条带在虹鳟三倍体群体中占比分别为62.50%、58.33%和70.83%,而在虹鳟二倍体群体中占比均为0%。使用所选的3个引物通过观察等位基因数目和3条特异等位基因条带来鉴定虹鳟样本的倍性(表 4)。
|
|
表 2 鉴别虹鳟三倍体的特异标记信息 Tab.2 Specific microsatellite markers for identification of triploid rainbow trout |
|
|
表 3 虹鳟二倍体、三倍体在3个标记中出现的比例差异 Tab.3 The differences in proportional distribution of the 3 microsatellite markers between diploid and triploid groups of rainbow trout |
|
图 2 引物SSR1054(a)、SSR1056(b)和SSR1468(c)扩增虹鳟基因组DNA的电泳 Fig.2 Electrophoretic patterns of primer SSR1054 (a), SSR1056 (b) and SSR1468 (c) in multiplication of genomic DNA in rainbow trout M:分子量标准DL1000;2n:虹鳟二倍体个体;3n:虹鳟三倍体个体 M: Molecular weight standard DL1000; 2n: Diploid individual rainbow trout; 3n: Individual triploid rainbow trout |
|
|
表 4 3个特异标记组合对三倍体虹鳟个体的鉴别结果 Tab.4 Identification results of three specific microsatellite markers for individual triploid rainbow trout |
用所选的3个鉴定虹鳟倍性水平的标记对48个未知倍性虹鳟样本进行PCR扩增,其中,扩增出现特异标记条带或3条条带的样本均为三倍体,其余个体判定为虹鳟二倍体。如表 5所示,SSR1054样本扩增图谱中含有三倍体特异等位基因305 bp或3条带基因型的个体判定为虹鳟三倍体个体,共检测出三倍体个体29个,SSR1056共检测出三倍体个体27个,SSR1468共检测出三倍体个体35个。结合特异标记SSR1054、SSR1056和SSR1468判定为虹鳟三倍体为46尾,与流式细胞仪检测倍性结果一致。
|
|
表 5 3个SSR特异标记在倍性鉴定中的有效性验证结果 Tab.5 Validation results of three specific microsatellite markers in ploidy identification |
本研究通过对153个虹鳟SSR标记筛选,结合计算和实验方法,最终选择3个特异SSR标记用于虹鳟三倍体鉴定。在实验过程中发现,这3个SSR引物在正规实验操作的情况下频繁出现失活的现象,因此,利用Primer3重新在SSR位点的侧翼区域设计引物对,最终得到3对稳定的引物序列。结果显示,所有52个虹鳟参考样本以及48个倍性未知样本的倍性水平均被正确区分开,这证实了本研究所筛选的特异SSR标记和相关分析方法的可靠性和实用性。
在选择用于倍性鉴定的SSR标记时,本研究优先选择具有高变异性的扩增位点,这将大大提高SSR辅助倍性鉴定的效率(Guo et al, 2016)。对扩增位点变异性的区分主要考虑表达的等位基因数量,以及这些等位基因的相对频率。此外,具有易于观察分析的清晰图谱条带也是筛选优质特异SSR标记的重要标准之一。因为通过统计清晰的图谱条带可以正确地判断每个位点上的等位基因构型,从而确定虹鳟的倍性水平。
利用SSR标记对多倍体物种进行群体遗传学研究是具有挑战性的,因为当电泳图谱中显示的微卫星DNA等位基因数量少于该多倍体物种倍性水平可能出现的最大等位基因数量时,很难通过直接观察等位基因数量来准确判断其倍性(Ferrante et al, 2010; Palop-Esteban et al, 2011)。在二倍体个体中,高度杂合的SSR在一个特定的位点上最多只能扩增2个等位基因。在三倍体和四倍体个体中,则可观察到3个和4个等位基因(Feng et al, 2018)。然而,在一些特殊情况下,无法通过直接观察等位基因数目来正确判断等位基因的实际数量。当只有1个独特等位基因时,无法准确判断其基因型。同理,倍性未知的个体显示2个等位基因数目也有多种可能的基因型,二倍体(AB)和三倍体(AAB或ABB)都可能发生这种情况(表 6)。本研究通过结合多个特异SSR标记的分析结果来提高倍性检测的准确性。如图 2b中三倍体的17号参考样本在图谱中仅显示1条条带,但在图 2a和图 2c中三倍体的17号样本显示3条条带。此外,本研究发现了3条可以辅助区分虹鳟二倍体和三倍体的特异标记条带,分别是SSR1054-305bp、SSR1056-463bp和SSR1468-363bp。这3条特异性标记条带将大大提升使用SSR标记鉴定虹鳟三倍体的准确率。
|
|
表 6 虹鳟二倍体和三倍体所有等位基因构型的预期电泳条带数 Tab.6 The expected number of electrophoretic bands of all possible allelic configurations of diploid and triploid rainbow trout |
微卫星无效等位基因是SSR在倍性鉴定中的另一个潜在的限制,它是由于SSR侧冀序列突变等导致基因无法正常扩增,它可以通过改变引物与侧翼序列的结合位点来消除或避免(Ellis et al, 2007; Hernández-Urcera et al, 2012; Nie et al, 2014)。在使用SSR标记倍性鉴定时,无效等位基因可能会导致实验统计的等位基因数目比实际等位基因数目低。在本研究中,筛选的3个特异SSR标记在分析的100个虹鳟样本过程中没有受到无效等位基因影响。这可能由于这些微卫星是从虹鳟物种本身基因组中开发的,如果将这3个SSR引物应用于近缘物种时,会提高无效等位基因的出现频率(Palop-Esteban et al, 2011)。此外,已有研究表明,当采用适当数量的具有高度可变性的SSR标记时,无效等位基因在倍性鉴定中不会产生严重影响,因为SSR位点的大量变异性补偿了由任何一个位点引入的潜在偏差影响(Biasi et al, 2015; Lemer et al, 2011)。虽然本研究的结果不能在未来的样本中完全摒弃无效等位基因的存在,但目前的实验数据表明,无效等位基因出现的频率非常低,它们对多倍体鉴定的影响将是微乎其微的。
综上所述,本研究所筛选出的一组3个虹鳟倍性鉴定特异SSR标记,为区分虹鳟二倍体、三倍体提供了一种微创、经济、可靠和可批量操作的分子方法。该方法可用于虹鳟三倍体制种过程对发眼卵、幼鱼等进行倍性监测的质控环节。
ALLEN S K. Flow cytometry: Assaying experimental polyploid fish and shellfish. Aquaculture, 1983, 33(1/2/3/4): 317-328 |
BAI Z, LIU F, LI J, et al. Identification of triploid individuals and clonal lines in Carassius auratus complex using microsatellites. International Journal of Biological Sciences, 2011, 7(3): 279-285 DOI:10.7150/ijbs.7.279 |
BIASI A, MARTIN F, SCHENA L. Identification and validation of polymorphic microsatellite loci for the analysis of Phytophthora nicotianae populations. Journal of Microbiological Methods, 2015, 110: 61-67 DOI:10.1016/j.mimet.2015.01.012 |
CLEVELAND B M, WEBER G M. Effects of steroid treatment on growth, nutrient partitioning, and expression of genes related to growth and nutrient metabolism in adult triploid rainbow trout (Oncorhynchus mykiss). Domestic Animal Endocrinology, 2016, 56: 1-12 DOI:10.1016/j.domaniend.2016.01.001 |
COULIBALY I, GHARBI K, DANZMANN R G, et al. Characterization and comparison of microsatellites derived from repeat-enriched libraries and expressed sequence tags. Animal Genetics, 2005, 36(4): 309-315 DOI:10.1111/j.1365-2052.2005.01305.x |
ELLIS J R, BURKE J M. EST-SSRs as a resource for population genetic analyses. Heredity, 2007, 99(2): 125-132 DOI:10.1038/sj.hdy.6801001 |
FENG B, YI S V, ZHANG M, et al. Development of novel EST-SSR markers for ploidy identification based on de novo transcriptome assembly for Misgurnus anguillicaudatus. PLoS One, 2018, 13(4): e195829 |
FERRANTE S P, LUCRETTI S, REALE S, et al. Assessment of the origin of new citrus tetraploid hybrids (2n=4X) by means of SSR markers and PCR based dosage effects. Euphytica, 2010, 173(2): 223-233 DOI:10.1007/s10681-009-0093-3 |
GLOVER K A, BOS J B, URDAL K, et al. Genetic screening of farmed Atlantic salmon escapees demonstrates that triploid fish display reduced migration to freshwater. Biological Invasions, 2016, 18(5): 1287-1294 DOI:10.1007/s10530-016-1066-9 |
GUO W, HOU J, YIN T, et al. An analytical toolkit for polyploid willow discrimination. Scientific Reports, 2016, 6: 37702 DOI:10.1038/srep37702 |
HAN Y, LIU M, LAN ZHANG L, et al. Comparison of reproductive development in triploid and diploid female rainbow trout Oncorhynchus mykiss. Journal of Fish Biology, 2010, 76(7): 1742-1750 DOI:10.1111/j.1095-8649.2010.02613.x |
HERNÁNDEZ-URCERA J, VERA M, MAGADÁN S, et al. Development and validation of a molecular tool for assessing triploidy in turbot (Scophthalmus maximus). Aquaculture, 2012, 330/331/332/333: 179-184 |
HUANG T, GU W, LIU E, et al. Comprehensive analysis of miRNA-mRNA/lncRNA during gonadal development of triploid female rainbow trout (Oncorhynchus mykiss). Genomics, 2021, 113(6): 3533-3543 DOI:10.1016/j.ygeno.2021.08.018 |
KARLSSON S, DISERUD O H, FISKE P, et al. Widespread genetic introgression of escaped farmed Atlantic salmon in wild salmon populations. ICES Journal of Marine Science, 2016, 73(10): 2488-2498 DOI:10.1093/icesjms/fsw121 |
KRISFALUSI M, CLOUD J G. Gonadal sex reversal in triploid rainbow trout. Journal of Experimental Zoology, 1999, 284(4): 466-472 DOI:10.1002/(SICI)1097-010X(19990901)284:4<466::AID-JEZ13>3.0.CO;2-G |
LAMPERT K P, LAMATSCH D K, SCHORIES S, et al. Microsatellites for the gynogenetic amazon molly, Poecilia formosa: Useful tools for detection of mutation rate, ploidy determination and overall genetic diversity. Journal of Genetics, 2006, 85(1): 67-71 DOI:10.1007/BF02728973 |
LECOMMANDEUR D, HAFFRAY P, PHILIPPE L. Rapid flow cytometry method for ploidy determination in salmonid eggs. Aquaculture Research, 1994, 25(3): 345-350 DOI:10.1111/j.1365-2109.1994.tb00698.x |
LEMER S, ROCHEL E, PLANES S. Correction method for null alleles in species with variable microsatellite flanking regions, a case study of the black-lipped pearl oyster Pinctada margaritifera. Journal of Heredity, 2011, 102(2): 243-246 DOI:10.1093/jhered/esq123 |
MANOR M L, WEBER G M, CLEVELAND B M, et al. Effects of feeding level and sexual maturation on fatty acid composition of energy stores in diploid and triploid rainbow trout (Oncorhynchus mykiss). Aquaculture, 2014, 418/419: 17-25 DOI:10.1016/j.aquaculture.2013.09.023 |
MANOR M L, WEBER G M, SALEM M, et al. Effect of sexual maturation and triploidy on chemical composition and fatty acid content of energy stores in female rainbow trout, Oncorhynchus mykiss. Aquaculture, 2012, 364/365: 312-321 DOI:10.1016/j.aquaculture.2012.08.012 |
MISHINA T, TAKADA M, TAKESHIMA H, et al. Molecular identification of species and ploidy of carassius fishes in Lake Biwa, using mtDNA and microsatellite multiplex PCRs. Ichthyological Research, 2014, 61(2): 169-175 DOI:10.1007/s10228-014-0388-9 |
NIE H, LI Q, KONG L, et al. Genotyping based on telomeric microsatellite loci for verifying triploidy in the Pacific oyster, Crassostrea gigas. Biochemical Systematics and Ecology, 2014, 54: 326-332 DOI:10.1016/j.bse.2014.03.007 |
NYNCA J, SŁOWIŃSKA M, WIŚNIEWSKA J, et al. Ovarian transcriptome analysis of diploid and triploid rainbow trout revealed new pathways related to gonadal development and fertility. Animal, 2022, 16(8): 100594 DOI:10.1016/j.animal.2022.100594 |
OHARA K, ARIYOSHI T, SUMIDA E, et al. Natural hybridization between diploid crucian carp species and genetic independence of triploid crucian carp elucidated by DNA markers. Zoological Science, 2000, 17(3): 357-364 |
OHARA K, DONG S, TANIGUCHI N. High proportion of heterozygotes in microsatellite DNA loci of wild clonal silver crucian carp, Carassius langsdorfii. Zoological Science, 1999, 16(6): 909-913 DOI:10.2108/zsj.16.909 |
PALOP-ESTEBAN M, SEGARRA-MORAGUES J G, GONZÁLEZ-CANDELAS F. Polyploid origin, genetic diversity and population structure in the tetraploid sea lavender Limonium narbonense Miller (Plumbaginaceae) from eastern Spain. Genetica, 2011, 139(10): 1309-1322 DOI:10.1007/s10709-012-9632-2 |
PHILLIPS R B, NICHOLS K M, DEKONING J J, et al. Assignment of rainbow trout linkage groups to specific chromosomes. Genetics, 2006, 174(3): 1661-1670 DOI:10.1534/genetics.105.055269 |
QI Y R, WANG Y P, HAO X L, et al. DNA flow cytometry and its application in the detection of plant genome size and ploidy. Molecular Plant Breeding, 2022, 20(7): 2279-2285 [齐嫣然, 王英平, 郝小丽, 等. DNA流式细胞术及其在植物基因组大小与倍性检测中的研究与应用. 分子植物育种, 2022, 20(7): 2279-2285] |
REXROAD C E 3rd, COLEMAN R L, GUSTAFSON A L, et al. Development of rainbow trout microsatellite markers from repeat enriched libraries. Marine Biotechnology, 2002a, 4(1): 12-16 DOI:10.1007/s10126-001-0058-6 |
REXROAD C E 3rd, COLEMAN R L, MARTIN A M. Thirty-five polymorphic microsatellite markers for rainbow trout (Oncorhynchus mykiss). Animal Genetics, 2001, 32(5): 317-319 DOI:10.1046/j.1365-2052.2001.0730b.x |
REXROAD C E 3rd, COLEMAN R L, HERSHBERGER W K, et al. Eighteen polymorphic microsatellite markers for rainbow trout (Oncorhynchus mykiss). Animal Genetics, 2002b, 33(1): 76-78 DOI:10.1046/j.1365-2052.2002.0742d.x |
REXROAD C E 3rd, COLEMAN R L, HERSHBERGER W K, et al. Rapid communication: Thirty-eight polymorphic microsatellite markers for mapping in rainbow trout. Journal of Animal Science, 2002c, 80(2): 541-542 DOI:10.2527/2002.802541x |
ROBINSON A J, LOVE C G, BATLEY J, et al. Simple sequence repeat marker loci discovery using SSR primer. Bioinformatics, 2004, 20(9): 1475-1476 DOI:10.1093/bioinformatics/bth104 |
SU Z G, XU K S, CHEN B P, et al. Studies on triploid silver carp and its karyotype. Zoological Research, 1984, 5(3): 15-22 [苏泽古, 许克圣, 陈尚萍, 等. 白鲢三倍体及其核型的研究. 动物学研究, 1984, 5(3): 15-22] |
WANG B Q, GU W, XU G F. Rainbow trout var. Shuike-1. China Fisheries, 2021(10): 89-95 [王炳谦, 谷伟, 徐革锋. 虹鳟"水科1号". 中国水产, 2021(10): 89-95] |
WERNER C, POONTAWEE K, MUELLER-BELECKE A, et al. Flesh characteristics of pan-size triploid and diploid rainbow trout (Oncorhynchus mykiss) reared in a commercial fish farm. Archiv für Tierzucht, 2008, 51(1): 71-83 |
XU L, ZHAO M, RYU J H, et al. Reproductive sterility in aquaculture: A review of induction methods and an emerging approach with application to Pacific northwest finfish species. Reviews in Aquaculture, 2022, 15(1): 220-241 |
YIN H B. Study on the karyotypes of four kinds carp. Chinese Journal of Fisheries, 2001(1): 7-10 [尹洪滨. 四种鲤鱼染色体核型比较研究. 水产学杂志, 2001(1): 7-10] |
YIN J, ZHAO Z S, CHEN X Q, et al. Karyotype comparison of diploid and tetraploid loach. Acta Hydrobiologica Sinica, 2005, 29(4): 469-472 [印杰, 赵振山, 陈小奇, 等. 二倍体和四倍体泥鳅染色体组型比较. 水生生物学报, 2005, 29(4): 469-472 DOI:10.3321/j.issn:1000-3207.2005.04.021] |



