2. 海水养殖生物育种与可持续产出全国重点实验室 中国水产科学研究院黄海水产研究所 山东 青岛 266071;
3. 青岛海洋科技中心海洋渔业科学与食物产出过程功能实验室 山东 青岛 266237;
4. 日照市海洋与渔业研究院 山东 日照 276800;
5. 江苏海洋大学 江苏 连云港 222005
2. State Key Laboratory of Mariculture Biobreeding and Sustainable Goods, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao 266071, China;
3. Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao Marine Science and Technology Center, Qingdao 266237, China;
4. Rizhao Ocean and Fishery Research Institute, Rizhao 276800, China;
5. Jiangsu Ocean University, Lianyungang 222005, China
热休克蛋白(heat shock proteins, HSPs),又称热应激蛋白,根据其分子质量,可分为HSP100、HSP90、HSP70、HSP60和小分子热休克蛋白(sHSPs) 5个主要家族(Jee, 2016)。Ritossa (1962)首次在果蝇(Drosophila melanogaster)中发现HSP,随着进一步研究,发现其广泛存在于从细菌到哺乳动物等体内(Baringou et al, 2016; Qiu et al, 2023; Salway et al, 2011; Wu et al, 2022)。研究发现,HSPs在早期胚胎发育和生殖细胞成熟过程中发挥重要作用,可能与生长发育过程中细胞的增殖、形态变化和细胞凋亡有关(Rupik et al, 2011; Wang et al, 2016)。Dmhsp70-A和Dmhsp70-B在大型蚤(Daphnia magna)无性生殖胚胎发育过程中表现出相似的变化趋势,在有性生殖胚胎发育过程中则完全不同,但均处于较高的表达水平(Chen et al, 2021);Li等(2022)利用RNAi技术研究hsp70对丰年虫(Artemia salina)幼体的保护作用,结果显示,与具有hsp70的幼体相比,敲除hsp70的幼体存活率显著下降;Takahashi等(2010)研究发现,hsp参与果蝇的发育;hsp70在黄条
日本对虾(Penaeus japonicus),又称日本囊对虾,其分布极广,在中国沿海、日本、东南亚及红海等均有分布,是中国、日本和东南亚等国家重要的商业虾种之一。据2023年中国渔业统计年鉴数据,我国日本对虾海水养殖面积达2.1万hm2,2022年日本对虾海水养殖总产量达4.6万t (农业农村部渔业渔政管理局, 2023)。已有相关研究表明,hsp在日本对虾中广泛存在,且在生长发育中发挥重要作用,如hsp10和hsp60在日本对虾各组织中均有表达且hsp60在早期发育阶段表达具有显著差异,进一步研究发现,HSP10与热休克因子(HSF)间存在互相作用(Zheng et al, 2018、2020a、2020b)。以上研究都是基于单个基因,但基于hsp家族在日本对虾生长发育中较为系统的研究尚未报道。目前,本实验室已完成日本对虾基因组测序工作(Ren et al, 2022),为解析hsp在日本对虾生长发育的分子机制提供了基础数据。
本研究拟通过生物信息学方法对日本对虾hsp基因家族进行鉴定和分析,并分析其在日本对虾发育时期的表达规律。通过生物信息学方法在日本对虾全基因组中鉴定出hsp基因家族的15个成员,分析它们的基因结构、基序组成、染色体定位和系统进化特征,并利用实时荧光定量PCR (qRT-PCR)技术研究日本对虾hsp基因家族在胚胎发育不同时期的表达规律,旨在为探讨hsp在日本对虾生长发育中的作用提供科学依据。
1 材料与方法 1.1 Pjhsp基因家族成员的鉴定及蛋白特征分析日本对虾基因组数据来自本实验室(Ren et al, 2022),同时在Ensembl和NCBI数据库中分别下载人(Homo sapiens)、狗(Canis lupus familiaris)、果蝇、小鼠(Mus musculus)、斑马鱼(Danio rerio)、凡纳滨对虾(Litopenaeus vannamei)、中国对虾(Fenneropenaeus chinensis)、斑节对虾(Penaeus monodon)、刀额新对虾(Metapenaeus ensis)、脊尾白虾(Exopalaemon carinicauda)、克氏原螯虾(Procambarus clarkia)、三疣梭子蟹(Portunus trituberculatus)和中华绒螯蟹(Eriocheir sinensis)已发表的HSP蛋白序列作为参考。使用TBtools软件(Chen et al, 2020)从日本对虾基因组中进行CDS序列提取并翻译为蛋白序列,使用TBtools软件的BLASTP程序对日本对虾基因组进行多序列比对,筛选出hsp基因家族成员。利用ExPASy (https://www.expasy.org/) (Gasteiger et al, 2005)网站的ProtParam程序预测HSP的分子量、等电点、不稳定系数和亲水性平均系数等分子特征,利用PSORT网站(https://psort.hgc.jp/form2.html)(Nakai et al, 2019)对HSP进行亚细胞定位。
1.2 PjHSP的系统发育分析使用MEGA 11 (Tamura et al, 2021)软件中的ClustalW分别对PjHSP和上述物种中已鉴定的HSP蛋白序列进行多序列比对,基于相邻连接法(neighbor-joining, NJ)构建系统发育树,bootstrap检测重复500次。使用在线网站ChiPlot (https://www.chiplot.online/index.html)(Xie et al, 2023)对发育树进行优化和展示。
1.3 Pjhsp基因特征分析为分析日本对虾hsp的基因结构,使用TBtools软件(Chen et al, 2020)从日本对虾基因组注释文件中提取hsp基因的结构信息,使用GSDS网站(http://gsds.gaHSPo-lab.org/)(Hu et al, 2015)绘制基因结构图。
使用MEME Suite网站(https://meme-suite.org/meme/tools/meme)(Bailey et al, 2015)对HSP蛋白序列的保守基序进行预测分析。设置每个序列0或1个基序,最大基序数为30个,最佳基序宽度为30~100个氨基酸。
使用NCBI Batch Web CD-Search Tool网站(https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (Marchler-Bauer et al, 2010)预测HSP保守结构域。使用TBtools软件对其进行可视化处理,并对Pjhsp家族成员进行染色体定位和可视化处理(Chen et al, 2020)。
1.4 样品采集实验用日本对虾亲虾来自台湾澎湖群体。挑选性腺发育Ⅳ期或以上亲虾培育产卵,取卵进行镜检和取样。日本对虾发育分期参照Hudinaga (1942)的研究,合子期(MZ)、囊胚期(B)、原肠期(G)、肢芽期(LB)和膜内无节期(IN)均用200目纱网过滤后取30颗卵,无节幼体Ⅰ期(N1)、溞状幼体Ⅰ期(Z1)、糠虾Ⅰ期(M1)和仔虾Ⅰ期(P1)均用80目纱网过滤后取15尾。所有样品置于2 mL冻存管后迅速放入液氮罐中,后转入–80 ℃冰箱保存,用于后期RNA的提取。
1.5 Pjhsp在日本对虾发育不同时期的表达分析采用Trizol法提取RNA,使用分光光度计和琼脂凝胶电泳检测RNA纯度和质量。使用HiScriptⅢ RT SuperMix for qPCR (+gDNA wiper)(诺唯赞)合成cDNA。利用Primer Premier 5.0软件设计引物(表 1),EF1-α为内参基因,引物由生工生物工程(上海)股份有限公司合成。使用ChamQ SYBR Color qPCMaster Mix (诺唯赞)进行qRT-PCR检测Pjhsp在日本对虾发育不同时期的表达量。反应体系20 μL:10 μL 2×ChamQ SYBR Color qPCR Master Mix、0.4 μL 50×ROX Reference Dye 1、0.4 μL上游引物、0.4 μL下游引物、2 μL模板和6.8 μL ddH2O。反应程序:95 ℃预变性30 s;95 ℃变性10 s,60 ℃退火30 s,循环40次;熔解曲线:95 ℃ 15 s,60 ℃ 60 s,95 ℃ 15 s。
|
|
表 1 实验所用引物序列 Tab.1 Sequence of primers used in this study |
采用2–ΔΔCT (Livak et al, 2001)方法计算基因相对表达量。结果用平均值±标准差(Mean±SD)表示。用SPSS 26.0进行单因素方差分析(one-way ANOVA)和Duncan多重比较,显著性水平设为P < 0.05。GraphPad Prism软件制作折线图。
2 结果 2.1 Pjhsp基因家族成员的鉴定及蛋白特征分析日本对虾基因组中共鉴定出15个hsp基因家族成员,根据保守结构域分析,对其进行初步分类,将其分为Pjhsp10、Pjhsp20、Pjhsp40、Pjhsp60、Pjhsp70和Pjhsp90 6个基因家族。各家族成员的基因ID、蛋白名称、蛋白序列长度和分子量大小等信息如表 2所示。PjHSP蛋白序列长度为102~795个氨基酸,分子量范围为10.95~91.48 kDa,其中,PjHSP90-25.279最长,为795 aa;PjHSP10-19.517最短,为102 aa。蛋白质等电点范围为4.54~9.33,其中,PjHSP20-1.821等电点最小,PjHSP70-15.369等电点最大。蛋白质不稳定系数大于40的包括PjHSP20-1.748、PjHSP20-1.821、PjHSP40-21.13、PjHSP70-15.369、PjHSP70-1.298、PjHSP90-12.759、PjHSP90-25.279和PjHSP90-31.274,其他小于40。蛋白疏水性计算分析表明,除了PjHSP10-19.517有疏水性,其他蛋白均有一定程度的亲水性。亚细胞定位结果显示,细胞质中有7个,细胞核中有2个,细胞外基质中有2个,内质网中有2个,线粒体中有2个。
|
|
表 2 Pjhsp基因家族蛋白质组成和理化性质 Tab.2 Protein composition and physicochemical properties of Pjhsp gene family |
基于日本对虾、凡纳滨对虾、斑节对虾、中国对虾、克氏原螯虾、刀额新对虾、脊尾白虾、三疣梭子蟹、中华绒螯蟹、菲律宾蛤仔(Ruditapes philippinarum)、黑腹果蝇、斑马鱼、小鼠、二化螟(Chilo suppressalis)、沙葱荧叶甲(Galeruca daurica)和麦红吸浆虫(Sitodiplosis mosellana)中已鉴定的HSP建立系统发育树(图 1)。发育树主要分为3大支,其中,HSP10、HSP20和HSP60家族聚为一支,HSP40和HSP90蛋白家族聚为一支,HSP70家族单独聚为一支。在HSP10家族中,PjHSP10-19.527与斑节对虾、中国对虾和凡纳滨对虾等HSP10聚为一支;在HSP20家族中,PjHSP20-1.748与PjHSP20-1.821单独聚为一支;在HSP40家族中,PjHSP40-21.13和PjHSP40-15.349单独聚为一支;在HSP60家族中,PjHSP60-19.518与中国对虾和凡纳滨对虾HSP60聚为一支;在HSP70家族中,PjHSP70-32.916与日本对虾HSP70聚为一支,然后与斑节对虾和脊尾白虾HSP70聚为一支,最后分别与PjHSP70-15.369、PjHSP70-3.662、PjHSP70-29.326、PjHSP70-39.287和PjHSP70-1.298聚为一大支;在HSP90家族中,PjHSP90-25.279先与凡纳滨对虾和斑马鱼HSP90聚为一支,然后与PjHSP90-12.729和PjHSP90-31.274聚为一支。
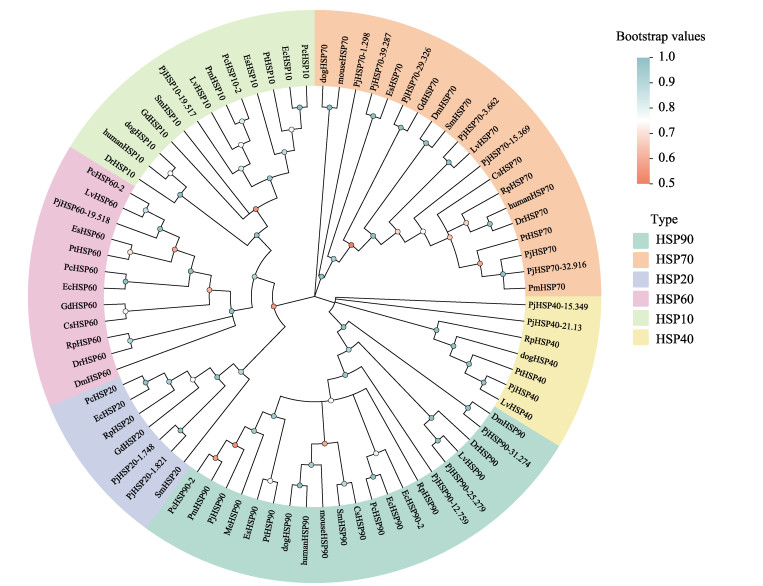
|
图 1 16个物种HSP系统发育树 Fig.1 Phylogenetic tree of HSP among sixteen species Pj:日本对虾;Lv:凡纳滨对虾;Pm:斑节对虾;Fc:中国对虾;Pc:克氏原螯虾;Me:刀额新对虾;Ec:脊尾白虾;Pt:三疣梭子蟹;Es:中华绒螯蟹;Rp:菲律宾蛤仔;Dm:黑腹果蝇;Dr:斑马鱼;mouse:小鼠;Cs:二化螟;Gd:沙葱荧叶甲;Sm:麦红吸浆虫;dog:狗;human:人。 Pj: Penaeus japonicus; Lv: Litopenaeus vannamei; Pm: Penaeus monodon; Fc: Fenneropenaeus chinensis; Pc: Procambarus clarkia; Me: Metapenaeus ensis; Ec: Exopalamon carincauda; Pt: Portunus trituberculatus; Es: Eriocheir sinensis; Rp: Ruditapes philippinarum; Dm: Drosophila melanogaster; Dr: Danio rerio; Mouse: Mus musculus; Cs: Chilo suppressalis; Gd: Galeruca daurica; Sm: Sitodiplosis mosellana; dog: Canis lupus familiaris; human: Homo sapiens. |
图 2所示为Pjhsp基因家族15个成员的基因结构示意图。不同的基因在染色体上长度不同,且内含子长度及数量均存在差异,其中,Pjhsp90-25.279内含子数量最多(14个),其次是PjhsP70-39.287,含13个内含子,Pjhsp70-15.369和Pjhsp70-32.916中未发现内含子。
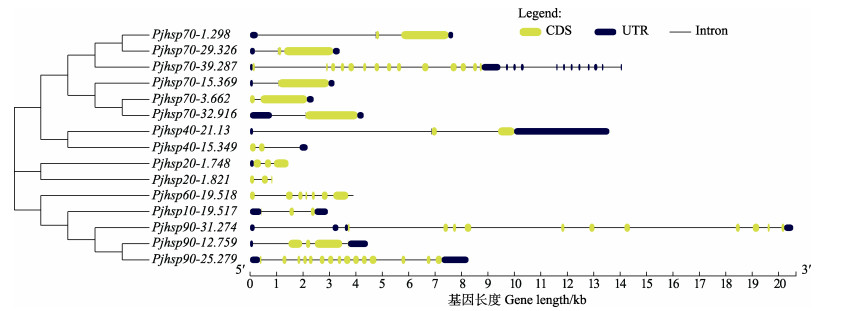
|
图 2 Pjhsp基因结构 Fig.2 Gene structure of Pjhsp gene family |
Motif分析显示(表 3和图 3),蛋白序列长度在30~100个氨基酸之间,同一HSP蛋白家族中motif排列顺序和数量相似,不同家族间motif差异较大。其中,motif 12、motif 15、motif 17、motif 25和motif 28分别由2个家族共有,其他motif均具有家族特异性。PjHSP10-19.517中未发现motif;PjHSP20家族和PjHSP40家族均有4个motif;PjHSP60家族有3个motif;PjHSP70家族和PjHSP90家族均有12个motif。
|
|
表 3 PjHSP家族蛋白基序信息 Tab.3 Protein motif information of PjHSP family |
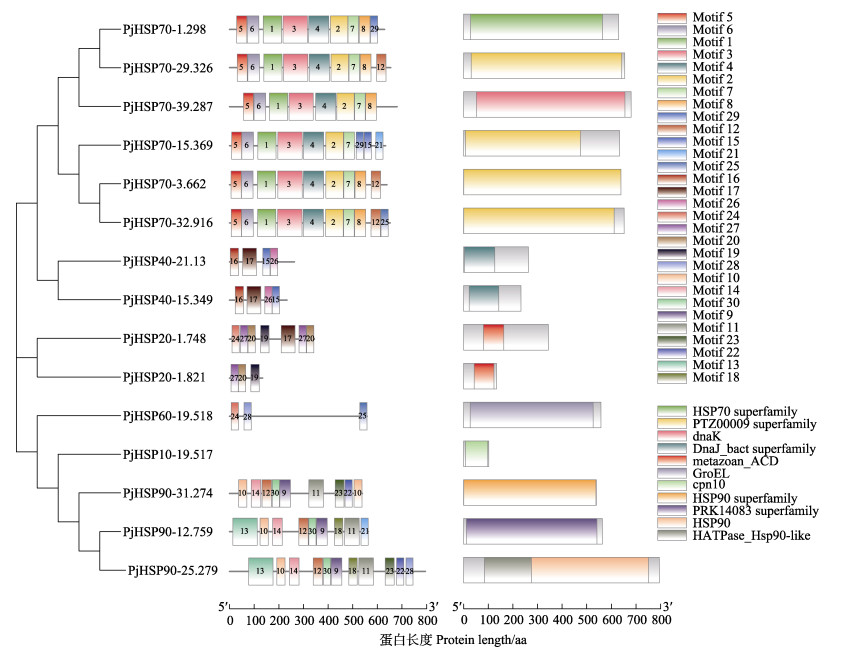
|
图 3 PjHSP家族蛋白基序和结构域 Fig.3 Motifs and domains of PjHSP family |
结构域分析显示(图 3),PjHSP10-19.517含有cpn10结构域;PjHSP20-1.748和PjHSP20-1.821含有metazoan ACD结构域;PjHSP40-21.13和PjHSP40-15.349含有DnaJ bact superfamily结构域;PjHSP60-19.518含有GroEL结构域;PjHSP70-15.369、PjHSP70-32.916、PjHSP70-3.662和PjHSP70-29.326含有PTZ00009 superfamily结构域,PjHSP70-39.287含有dnaK结构域,PjHSP70-1.298含有HSP70 superfamily结构域;PjHSP90-31.274含有HSP90 superfamily结构域,PjHSP90-12.759含有PRK14083 superfamily结构域,PjHSP90-25.279含有HATpase_HSP90-like结构域和HSP90结构域。
2.4 染色体定位Pjhsp基因家族的15个基因定位在11条染色体上(图 4),其中,Pjhsp70-1.298、Pjhsp20-1.748和Pjhsp20-1.821定位在第1条染色体上;Pjhsp70-3.662定位在第3条染色体上;Pjhsp90-12.759定位在第12条染色体上;Pjhsp40-15.349和Pjhsp70-15.369定位在第15条染色体上;Pjhsp10-19.517定位在第19条染色体上;Pjhsp40-21.13定位在第21条染色体上;Pjhsp60-19.518定位在第19条染色体上;Pjhsp90-25.279定位在第25条染色体上;Pjhsp70-29.326定位在第29条染色体上;Pjhsp90-31.274定位在第31条染色体上;Pjhsp70-32.916定位在第32条染色体上;Pjhsp70-39.287定位在第39条染色体上。
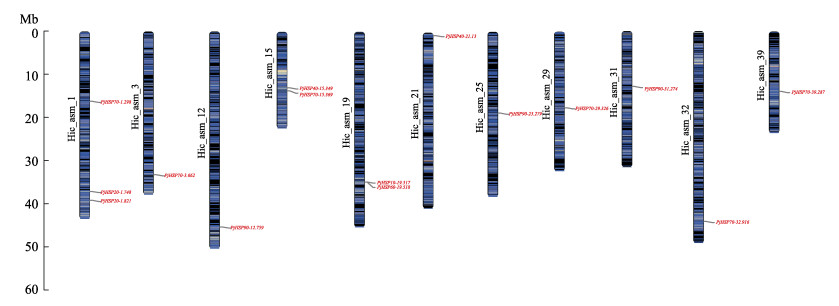
|
图 4 Pjhsp基因家族染色体分布 Fig.4 Chromosome location of Pjhsp gene family |
Pjhsp在胚胎发育所有时期均有表达。Pjhsp10-19.517在胚胎发育过程中的表达水平如图 5A所示,其在合子期至囊胚期表达显著下调(P < 0.05),囊胚期至仔虾Ⅰ期表达水平相对稳定,在合子期达到峰值,仔虾Ⅰ期最低。
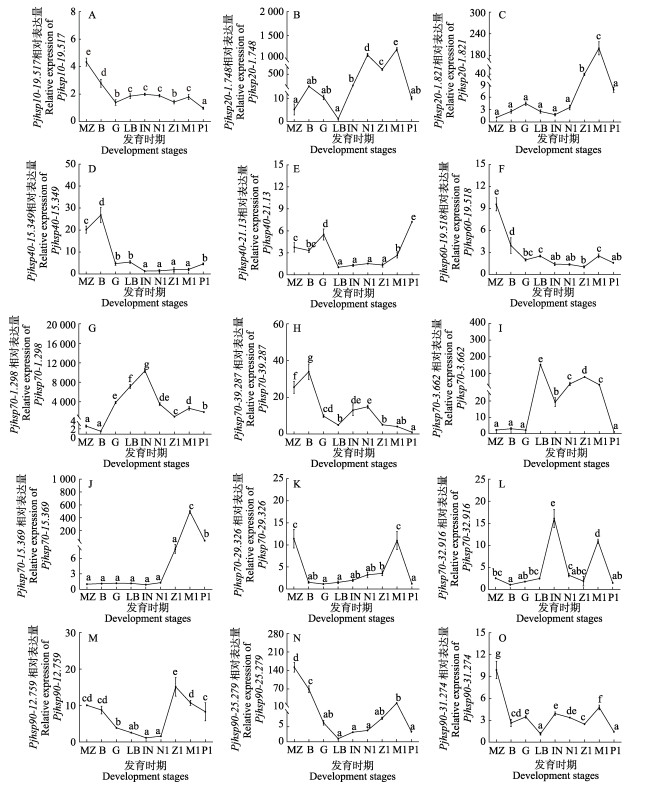
|
图 5 Pjhsp在日本对虾不同发育时期表达水平 Fig.5 Expression of Pjhsp during development stages of P. japonicus MZ:合子期;B:囊胚期;G:原肠期;LB:肢芽期;IN:膜内无节幼体期;N1:无节幼体Ⅰ期;Z1:溞状幼体Ⅰ期;M1:糠虾Ⅰ期;P1:仔虾Ⅰ期。不同字母表示组间差异显著(P < 0.05)。下同。 MZ: Maternal to zygotic; B: Blastocyst; G: Gastrula; LB: Limb buds; IN: Intramebranous nauplius; N1: Stage 1 of naulius; Z1: Stage 1 of zoea; M1: Stage 1 of mysis; P1: Stage 1 of post larvae. Significant differences between groups at P < 0.05 are indicated by different letters. The same below. |
Pjhsp20-1.748在胚胎发育过程中的表达水平如图 5B所示,其表达量在肢芽期最低且在其之前时期表达水平无显著变化(P > 0.05),随后显著上调(P < 0.05),在糠虾Ⅰ期达到峰值。Pjhsp20-1.821在胚胎发育过程中表达水平如图 5C所示,其在合子期表达量最低,直至无节幼体Ⅰ期之前表达相对稳定,无显著性变化(P > 0.05),随后表达显著上调(P < 0.05),在糠虾Ⅰ期达到峰值。
Pjhsp40-15.349在胚胎发育过程中的表达水平如图 5D所示,其表达水平在发育前期变化较显著,与合子期相比,囊胚期表达显著上调(P < 0.05),且在囊胚期达到峰值,随后显著下调(P > 0.05),在发育后期表达水平相对稳定。Pjhsp40-21.13在胚胎发育过程中的表达水平如图 5E所示,其表达水平在合子期和囊胚期无显著差异(P > 0.05);在原肠期比囊胚期的表达显著上调(P < 0.05);在肢芽期、膜内无节期、无节幼体Ⅰ期和溞状幼体Ⅰ期较稳定,无显著差异(P > 0.05),随后其表达显著上调,在仔虾Ⅰ期达到最高。
Pjhsp60-19.518在胚胎发育过程中的表达水平如图 5F所示,其表达水平合子期达到峰值,在合子期至原肠期显著下调(P < 0.05),随后至仔虾Ⅰ期表达水平相对稳定,在溞状幼体Ⅰ期达到最低。
Pjhsp70-1.298和Pjhsp70-3.662在胚胎发育过程中的表达水平如图 5G、I所示,整体变化趋势相似,均在发育中后期大量表达。其中,Pjhsp70-1.298表达水平在膜内无节期达到峰值,在囊胚期最低;Pjhsp70-3.662在肢芽期达到最高,在仔虾Ⅰ期最低。Pjhsp70-39.287和Pjhsp70-32.916在胚胎发育过程中的表达水平如图 5H、K所示,表达水平变化趋势较相似,不同的是,Pjhsp70-39.287在发育前期表达水平较高,在囊胚期达到最高,Pjhsp70-32.916在发育中后期表达较高,在膜内无节期达到最高;Pjhsp70-15.369在胚胎发育过程中的表达水平如图 5J所示,在合子期至无节幼体Ⅰ期表达无显著差异(P > 0.05),随后显著上调(P < 0.05),在糠虾Ⅰ期达到峰值;Pjhsp70-29.326在胚胎发育过程中表达水平如图 5L所示,在合子期表达最高,随后显著下调(P < 0.05),在囊胚期至溞状幼体Ⅰ期无显著变化(P > 0.05),随后显著上调(P < 0.05),在糠虾Ⅰ期达到较高表达水平,最后在仔虾Ⅰ期显著下调(P < 0.05)。
Pjhsp90-12.759和Pjhsp90-25.279在胚胎发育过程中表达水平如图 5M、N所示,整体变化趋势相似,从合子期表达下调(P < 0.05),其中,Pjhsp90-25.279在该时期表达最高,随后分别在膜内无节期和肢芽期降至最低,而后显著上调(P < 0.05),Pjhsp90-12.759在溞状幼体Ⅰ期达到最高;Pjhsp90-31.274在胚胎发育过程中的表达水平如图 5O所示,在合子期表达最高,随后显著下调(P < 0.05),小范围内波动变化,在肢芽期表达最低。
3 讨论 3.1 Pjhsp基因家族成员鉴定及特征分析hsp基因家族在亚洲长角甲虫(Anoplophora glabripennis)(Xu et al, 2022)、猛水蚤(Harpacticoid)和剑水蚤(Cyclopoid) (Park et al, 2021)等中已经报道。在甲壳动物中多数是基于单个hsp的基因特征分析及其在环境胁迫下的表达分析,研究hsp基因家族的相关报道较少(Sergio et al, 2020; Wisarut et al, 2021; Kifayatullah et al, 2023)。本研究在日本对虾基因组中共鉴定出15个Pjhsp基因家族成员,包括3个hsp90、6个hsp70、1个hsp60、2个hsp40、2个hsp20和1个hsp10家族成员。
等电点与蛋白本身所含酸性氨基酸和碱性氨基酸的数量有关,不同蛋白质因其氨基酸残基组成不同,等电点也不同。本研究中,多数PjHSP等电点 < 7,证明其可能为酸类蛋白(吴斌, 2003; 吴松锋, 2005)。亚细胞定位发现,PjHSP多数存在于细胞质中,但在细胞区域如内质网、线粒体和细胞核中也存在,相似的结果在马氏沼虾(Macrobrachium malcolmsonii) (Muthuswamy et al, 2018)、黄鳝(Monopterus albus) (杨文林等, 2023)和多种十足类(Baringou et al, 2016) hsp基因家族分析中有报道。多数PjHSP首先与斑节对虾、凡纳滨对虾和中国对虾等HSP聚类,之后与其他物种HSP聚类,与传统分类地位关系一致。Motif分析表明,在HSP90、HSP70、HSP40和HSP20蛋白家族中存在相同的motif,例如,HSP90家族共有motif 9、motif 10、motif 11、motif 12、motif 14和motif 30。说明hsp基因家族具有高度保守性,但在一些家族中无motif存在,如HSP10家族,因此,不同的家族间功能存在较大差异。
研究表明,一些结构域在HSPs分子伴侣方面发挥重要作用。例如cpn10结构域,其可与ATP结合,且cpn10结构域可自我组装形成圆柱形复合物,把错误折叠的蛋白隔离在中间,在大分子HSPs下进行折叠(Lissin et al, 1992);metazoan ACD结构域,其与多个亚基可形成具有活性的低聚复合物,以不依赖ATP的形式与错误折叠的蛋白质相结合,将其隔离在低聚复合物中,并通过依赖ATP型大分子HSP促进错误折叠蛋白的重新折叠,保证其空间结构的稳定性(Liu et al, 2015);DnaJ bact superfamily (HSP40)结构域,通过J结构域可与HSP70结合,作为辅助伴侣分子完成蛋白的正确折叠(何颖慧等, 2022);GroEL结构域可与其共伴侣分子HSP10形成一个中间具有空腔的复合物,为蛋白质的重新折叠提供空间(付青贤等, 2019)。本研究中,PjHSP蛋白家族均含有相应的结构域,表明HSP在分子伴侣方面发挥重要作用。
3.2 Pjhsp在日本对虾发育时期的表达分析研究表明,hsp在生物体发育过程中广泛表达,并表现出复杂的变化趋势。Zheng等(2018)研究发现,hsp60在日本对虾早期发育时期的表达具有显著差异。Iryani等(2020)研究发现,hsp70在丰年虫幼体发育过程中保护机体免受环境应激。Yang等(2019)研究发现,hsp在烟草甲(Lasioderma serricorne)幼虫和成虫期间的表达具有显著差异。本研究中,15个Pjhsp在合子期至仔虾Ⅰ期均有表达,表明hsp在日本对虾的早期发育中发挥重要作用。此外,不同基因家族变化趋势也不尽相同,如Pjhsp10-19.517与Pjhsp60-19.518的变化趋势相似,在胚胎发育时期大量表达,幼体发育时期显著下调,与Zheng等(2018)在日本对虾中hsp60在不同发育时期的表达的结果相似。相关研究表明,HSP60与HSP10互为分子伴侣,可完成对目的蛋白的正确折叠(Okamoto et al, 2015),这与本研究中Pjhsp60-19.518及Pjhsp10-19.517发育时期中的变化趋势一致。
Pjhsp在日本对虾发育时期的变化趋势可能因为如下原因。首先,日本对虾在胚胎发育过程中经历变态,该过程需要大量合成蛋白质,HSP作为分子伴侣参与蛋白的组装与正确折叠等过程。Pjhsp40、Pjhsp70-39.287和Pjhsp70-1.298等基因在胚胎时期大量表达,满足胚胎对大量蛋白质的需求。其次,Liang等(2020)发现,脊尾白虾中hsp90通过调控卵黄蛋白原的转录,间接调控胚胎发育过程中营养物质的供给,在胚胎发育过程中发挥重要作用。卵黄蛋白原作为卵黄蛋白的前体,几乎存在于所有卵生生物中,为胚胎发育提供营养(Pan et al, 2019)。本研究中,hsp90基因家族表达水平在合子期至肢芽期期间相对较高,推测hsp90基因家族在日本对虾胚胎发育时期显著上调表达,通过调控卵黄蛋白原的转录,满足胚胎对大量卵黄蛋白的需求,因此,日本对虾幼体表现出比胚胎时期更高的hsp90表达水平。最后,据报道,幼体时期与胚胎时期相比,其对环境应激更敏感(Lavarías et al, 2022)。因此,幼体易受水体环境影响造成机体应激,从而上调hsp表达,保护机体免受环境应激的危害;此外,日本对虾自无节幼体之后由内源性营养转变为外源性进食,而后者易受到病原体和异生素的影响,因此,Pjhsp90-12.759、Pjhsp70-3.662、Pjhsp70-15.369、Pjhsp70-32.916和Pjhsp20等在幼体时期上调表达可以保护幼体免受病原体和外源毒物的危害(Dai et al, 2020; Kumar et al, 2022)。
4 结论综上所述,从日本对虾基因组中共鉴定出15个hsp,其中7个HSP蛋白理化性质较稳定,不稳定系数小于40;亚细胞定位结果显示,7个HSP蛋白定位到细胞质,其余分别定位在细胞核、细胞外基质、内质网和线粒体。通过qRT-PCR技术分析Pjhsp在发育不同时期的表达规律,结果表明,在日本对虾发育不同时期hsp均有表达,推测其在信号传导和调节细胞周期中发挥重要作用;部分hsp如Pjhsp40、Pjhsp70-39.287和Pjhsp70-1.298等基因在胚胎时期大量表达,推测其通过发挥分子伴侣作用满足胚胎对大量蛋白质的需求并调控卵黄蛋白原的转录满足胚胎对卵黄蛋白的需求;部分hsp如Pjhsp90-12.759和Pjhsp70-3.662等基因在幼体时期大量表达,推测其通过协同免疫作用保护机体免受环境压力和病原生物等的侵害。
BAILEY T L, JOHNSON J, GRANT C E, et al. The MEME suite. Nucleic Acids Research, 2015, 43(1): 39-49 |
BARINGOU S, ROUAULT J D, KOKEN M, et al. Diversity of cytosolic HSP70 heat shock protein from decapods and their phylogenetic placement within Arthropoda. Gene, 2016, 591(1): 97-107 DOI:10.1016/j.gene.2016.06.061 |
Bureau of Fisheries Ministry of Agriculture and Rural Affairs, National Fisheries Technology Extension Center, China Society of Fisheries. China fishery statistical yearbook. Beijing: China Agriculture Press, 2022 [农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会. 中国渔业统计年鉴. 北京: 中国农业出版社, 2022]
|
CHEN C, CHEN H, ZHANG Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8): 1194-1202 DOI:10.1016/j.molp.2020.06.009 |
CHEN L, GOMEZ R, WEISS L C. Distinct gene expression patterns of two heat shock protein 70 members during development, diapause, and temperature stress in the freshwater crustacean Daphnia magna. Frontiers in Cell and Developmental Biology, 2021, 9: e692517 DOI:10.3389/fcell.2021.692517 |
DAI L S, KAUSAR S, GUL I, et al. Molecular characterization of a heat shock protein 21 (HSP21) from red swamp crayfish, Procambarus clarkii in response to immune stimulation. Developmental and Comparative Immunology, 2020, 111: e103755 DOI:10.1016/j.dci.2020.103755 |
FANG L, XU Y J, CUI A J, et al. Molecular cloning and temporal expression pattern of hsp70 gene during the early life stages of Seriola aureovittata. Progress in Fishery Sciences, 2023, 44(4): 55-63 [方璐, 徐永江, 崔爱君, 等. 黄条  hsp70基因克隆及其在早期生长发育过程中的表达调控特性. 渔业科学进展, 2023, 44(4): 55-63] hsp70基因克隆及其在早期生长发育过程中的表达调控特性. 渔业科学进展, 2023, 44(4): 55-63] |
FU Q X, YE Q Y. Research progress of heat shock protein 60 in nervous system diseases. Journal of International Neurology and Neurosurgery, 2019, 46(3): 341-345 [付青贤, 叶钦勇. 热休克蛋白60在神经系统疾病作用的研究进展. 国际神经病学神经外科学杂志, 2019, 46(3): 341-345] |
GASTEIGER E, HOOGLAND C, GATTIKER A, et al. Protein identification and analysis tools on the ExPASy server. Humana Press, 2005: 571-607
|
HE Y H, WANG Z P. Molecular chaperone HSP40/DNAJ protein family and its role in neurodegenerative diseases. Journal of Zhejiang University (Medical Sciences), 2022, 51(5): 640-646 [何颖慧, 王志萍. 分子伴侣HSP40/DNAJ蛋白家族及其在神经退行性疾病中的作用. 浙江大学学报(医学版), 2022, 51(5): 640-646] |
HU B, JIN J, GUO A Y, et al. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics (Oxford, England), 2015, 31(8): 1296-1297 |
HUDINAGA M. Reproduction, development and rearing of Penaeus japonicus bate. Journal of Zoology, 1942, 20(1): 305-389 |
IRYANI M T M, MACRAE T H, SORGELOOS P, et al. RNA interference of HSP70 in Artemia franciscana nauplii and its effect on morphology, growth, survival and immune response. Aquaculture, 2020, 520: e735012 DOI:10.1016/j.aquaculture.2020.735012 |
JEE H. Size dependent classification of heat shock proteins: A mini-review. Journal of Exercise Rehabilitation, 2016, 12(4): 255-259 DOI:10.12965/jer.1632642.321 |
KIFAYATULLAH M, GOLARA K, PAVEL K, et al. Heat shock proteins adaptive responses to environmental stressors and implications in health management of decapods. Aquaculture, 2023, 30: e101564 |
KUMAR V, ROY S, BEHERA BK, et al. Heat shock proteins (HSPs) in cellular homeostasis: A promising tool for health management in crustacean aquaculture. Life (Basel), 2022, 12(11): e1777 |
LAVARÍAS S M L, ARRIGHETTI F, LANDRO S M, et al. Sensitivity of embryos and larvae of the freshwater prawn Macrobrachium borellii to the latest generation pesticide spirotetramat. Ecotoxicology and Environmental Safety, 2022, 248: e114257 DOI:10.1016/j.ecoenv.2022.114257 |
LI Q, IRYANI M T M, LV A, et al. Effects of heat shock protein 70 knockdown on the tolerance of the brine shrimp Artemia franciscana to aquaculture-related stressors: Implications for aquatic animal health and production. Aquaculture, 2022, 550: e737872 DOI:10.1016/j.aquaculture.2021.737872 |
LIANG J P, ZHANG W L, LI H, et al. Abundances of vitellogenin and heat shock protein 90 during ovarian and embryonic development of Exopalaemon carinicauda. Animal Reproduction Science, 2020, 223: e106633 DOI:10.1016/j.anireprosci.2020.106633 |
LISSIN N M, SEDELNIKOVA S E, RYAZANTSEV S N. Crystallization of the cpn60/cpn10 complex ('holo-chaperonin') from Thermus thermophilus. FEBS Letters, 1992, 311(1): 22-24 DOI:10.1016/0014-5793(92)81357-R |
LIU L, CHEN J, YANG B, et al. Crystal structure and function of an unusual dimeric HSP20.1 provide insight into the thermal protection mechanism of small heat shock proteins. Biochemical and Biophysical Research Communications, 2015, 458(2): 429-434 DOI:10.1016/j.bbrc.2015.01.134 |
LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real–time quantitative PCR and the 2–ΔΔCT method. Methods, 2001, 25(4): 402-408 DOI:10.1006/meth.2001.1262 |
MARCHLER-BAUER A, LU S, ANDERSON J B, et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Research, 2010, 39(1): 225-229 |
MUTHUSWAMY K, SHANMUGAM PREMA D, KRISHNAN V, et al. Differential intracellular localization of HSP70 in the gill and heart tissue of fresh water prawn Macrobrachium malcolmsonii during thermal stress. Molecular Biology Reports, 2018, 45: 1321-1329 DOI:10.1007/s11033-018-4291-8 |
NAKAI K, IMAI K. Prediction of protein localization encyclopedia of bioinformatics and computational biology. Oxford: Academic Press, 2019: 53-59
|
OKAMOTO T, ISHIDA R, YAMAMOTO H, et al. Functional structure and physiological functions of mammalian wild-type HSP60. Archives of Biochemistry and Biophysics, 2015, 586: 10-19 DOI:10.1016/j.abb.2015.09.022 |
PAN X, LIU Y, ZHOU K, et al. Tissue expression and bioinformatics analysis of the vitellogenin gene of Asian arowana (Scleropages formosus). Journal of Applied Ichthyology, 2019, 35: 970-977 |
PARK J C, LEE J S. Genome-wide identification of heat shock proteins in harpacticoid, cyclopoid, and calanoid copepods: Potential application in marine ecotoxicology. Marine Pollution Bulletin, 2021, 169: e112545 DOI:10.1016/j.marpolbul.2021.112545 |
QIU Y, OZTURK S, CUI X, et al. Increased heat tolerance and transcriptome analysis of Salmonella enterica Enteritidis PT 30 heat-shocked at 42 ℃. Food Research International, 2023, 167: e112636 DOI:10.1016/j.foodres.2023.112636 |
REN X, LÜ J, LIU M, et al. A chromosome-level genome of the Kuruma shrimp (Marsupenaeus japonicus) provides insights into its evolution and cold–resistance mechanism. Genomics, 2022, 114(3): e110373 |
RITOSSA F. A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia, 1962, 18: 571-573 |
RUPIK W, JASIK K, BEMBENEK J, et al. The expression patterns of heat shock genes and proteins and their role during vertebrate's development. Comparative Biochemistry and Physiology Part A: Molecular and Integrative Physiology, 2011, 159(4): 349-366 |
SALWAY K D, GALLAGHER E J, PAGE M M, et al. Higher levels of heat shock proteins in longer-lived mammals and birds. Mechanisms of Ageing and Development, 2011, 132(6): 287-297 |
SERGIO A U, SALVADOR E L, MARÍA T S, et al. Litopenaeus vannamei oxygen consumption and HSP gene expression at cyclic conditions of hyperthermia and hypoxia. Journal of Thermal Biology, 2020, 92: e102666 |
TAKAHASHI K H, RAKO L, TAKANO-SHIMIZU T, et al. Effects of small HSP genes on developmental stability and microenvironmental canalization. Bmc Evolutionary Biology, 2010, 10: e284 |
TAMURA K, STECHER G, KUMAR S. MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 2021, 38(7): 3022-3027 |
WANG P F, ZENG S, XU P, et al. Two HSP90 genes in mandarin fish Siniperca chuatsi: Identification, characterization and their specific expression profiles during embryogenesis and under stresses. Fish Physiology and Biochemistry, 2016, 42(4): 1123-1136 |
WANG P, XU P, ZENG S, et al. Comparative analysis of sequence feature and expression of two heat shock cognate 70 genes in mandarin fish Siniperca chuatsi. Gene, 2015, 560(2): 226-236 |
WISARUT J, PREMRUETHAI S, ANCHALEE T. Structure, gene expression, and putative functions of crustacean heat shock proteins in innate immunity. Developmental and Comparative Immunology, 2021, 115: e103875 |
WU B. Study of enzyme properties in immobilized cells and molecular accessibility of protein. Master´s Thesis of Wuhan University, 2003 [吴斌. 固定化细胞酶性质及蛋白质分子可及性研究. 武汉大学硕士研究生学位论文, 2003]
|
WU J, GAO T, HU J, et al. Research advances in function and regulation mechanisms of plant small heat shock proteins (sHSPs) under environmental stresses. Science of the Total Environment, 2022, 825: e154054 |
WU S F. Basic bioinformatic studies of proteomic expression profile and multi-modality distribution of pI values in whole proteome. Doctoral Dissertation of Chinese People's Liberation Army Academy of Military Medical Science, 2005 [吴松锋. 蛋白质组表达谱基本生物信息学研究及全蛋白质组等电点分布研究. 中国人民解放军军事医学科学院博士研究生学位论文, 2005]
|
XIE J, CHEN Y, CAI G, et al. Tree visualization by one table (TBtools): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Research, 2023, 51(1): 587-592 |
XU Y B, SHI F M, LI Y R, et al. Genome-wide identification and expression analysis of the HSP gene superfamily in Asian long-horned beetle (Anoplophora glabripennis). International Journal of Biological Macromolecules, 2021, 200: 583-592 |
YANG W J, XU K K, CAO Y, et al. Identification and expression analysis of four small heat shock protein genes in cigarette beetle, Lasioderma serricorne (Fabricius). Insects, 2019, 10(5): e139 |
YANG W L, WEI H B, YANG D Q. Bioinformatics analysis of physical and chemical properties and molecular structure of Monopterus albus HSP70. Northern Chinese Fisheries, 2023, 42(4): 247-253 [杨文林, 魏红波, 杨代勤. 黄鳝HSP70理化性质和分子结构的生物信息学分析. 黑龙江水产, 2023, 42(4): 247-253] |
ZHENG J B, LI L, DONG H, et al. Molecular cloning of heat shock protein 60 from Marsupenaeus japonicus and its expression profiles at early developmental stages and response to heat stress. Aquaculture, 2018, 49: 301-312 |
ZHENG J B, MAO Y, SU Y, et al. Cross talk between heat shock protein 10 and a heat shock factor identified from Marsupenaeus japonicus. International Journal of Biological Macromolecules, 2020a, 147: 1041-1052 |
ZHENG J B, WANG P, MAO Y, et al. Full-length transcriptome analysis provides new insights into the innate immune system of Marsupenaeus japonicus. Fish and Shellfish Immunology, 2020b, 106: 283-295 |
ZHOU D, SU Y L, ZHONG R Z, et al. Molecular cloning and expression analysis of HSP90 of peanut worm Sipunculus nudus. Progress in Fishery Sciences, 2020, 41(5): 150-159 [周丹, 苏泳霖, 钟如卓, 等. 光裸星虫Hsp90基因的全长克隆及其在全组织和卵母细胞中的表达分析. 渔业科学进展, 2020, 41(5): 150-159] |



