2. Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao 266071;
3. School of Chemistry and Pharmaceutical Sciences, Qingdao Agriculture University, Qingdao 266109
Nucleic acid amplification is increasingly used in a broad array of applications, such as molecular biology, clinical diagnostics, food safety, and environmental monitoring, to name a few. In particular, it is still a gold standard technique for analyzing samples with a small amount of nucleotides (Goda et al, 2015; Stedtfeld et al, 2014). Although polymerase chain reaction (PCR) is the first and remains the most popular amplification technology in various fields, it suffers from several drawbacks such as the requirement of multiple thermo- cycling steps, easy contamination, and high cost, which largely limit its application in resource-limited settings and, specially, in point-of-care use (Zhao et al, 2015). To address this issue, isothermal nucleic acid amplification techniques have recently been developed. In particular, loop-mediated isothermal amplification (LAMP), which has the potential to revolutionize molecular biology by reducing the need for highly sophisticated equipment, and by having low running costs and short turnaround times, is in bloom (Zhao et al, 2015; Zhang et al, 2014). LAMP shows higher specificity than PCR because it uses four to six different primers that bind to specific sites on the template strand. Moreover, the sensitivity of LAMP is less affected by substances that usually inhibit PCR reactions, such as food ingredients and blood components (Zhang et al, 2014; Kaneko et al, 2007; Abdul-Ghani et al, 2012; Kiddle et al, 2012; Wang et al, 2008). This suggests that simple assays could be developed using LAMP with the most cumbersome steps of sample pretreatment, such as DNA extraction and purification, eliminated (Safavieh et al, 2016; Njiru et al, 2012; Williams et al, 2017). Up to now, several studies have been reported on its capacity to directly amplify target genes from rapidly processed, crude sample matrix (Poon et al, 2006; Njiru et al, 2008; Priye et al, 2017; Bektas et al, 2016; Hayashida et al, 2015; Lee et al, 2016; Koizumi et al, 2012; Soejima et al, 2011; Ihira et al, 2010), and even original samples with or without simple mechanical-based pretreatment (Williams et al, 2017; Hill et al, 2008; Patterson et al, 2013; Enomoto et al, 2005; Kanitkar et al, 2017; Stedtfeld et al, 2016; Youn et al, 2016; Williams et al, 2017; Ahmad et al, 2017), considerably reducing the cost and turnaround time.
Target genes can be directly detected by employing LAMP assays without involving the steps of DNA extraction and purification. For the success two key contributions should be acknowledged. First, the LAMP assay is tolerant of inhibition from complex substances in reaction matrices (Stedtfeld et al, 2014; Priye et al, 2017; Lee et al, 2016; Ahmad et al, 2017). Second, the acquired template plays a critical role. Some physical, chemical and biochemical methods, such as heat (Poon et al, 2006; Koizumi et al, 2012; Ihira et al, 2010), alkaline treatment (Bektas et al, 2016; Soejima et al, 2011) and addition of lysozyme (Lee et al, 2016), are sufficient for preparing DNA templates for LAMP. Furthermore, it has been reported that in some cases samples can be directly used as templates. This provides promise for simplifying the entire analysis operation. For example, Hill et al (2008) and Patterson et al (2013) found that unprocessed urine and whole blood could be added into LAMP reaction mixtures for the detection of target genes of specific bacteria. And in some other reports, the working mechanism has even been discussed preliminarily. Enomoto et al(2005) hypothesized that direct detection of HSV from swab samples originated from the large quantity of naked viral DNA as well as complete virions. Stedtfeld et al speculated that adequate cell lysis occurred while incubating LAMP reactions at 63℃ (Kanitkar et al, 2017; Stedtfeld et al, 2016). They also found that heat treatment prior to incubation yields comparable levels of sensitivity to those of direct amplification without lysis. Youn et al (2016) postulated that target genes in dead and viable bacterial cells could both be amplified, and Williams et al (2017) postulated that extracellular DNA, larger cells and particulates all contributed to reactions when a suspension of crushed veliger was used as a template. However, to the best of our knowledge, until now there are no dedicated reports on the validation of working templates and the behavior of viable cells in the LAMP reaction.
Since the end of last century, novel strategies based on the identification of DNA from environmental samples have proven noteworthy in detecting and monitoring not only common species, but also those that are endangered, invasive, or elusive (Williams et al, 2017; Ahmad et al, 2017; Bohmann et al, 2014; Giovannoni et al, 1990; Stoeck et al, 2010; Zielińska et al, 2017; Lee et al, 2017). In particular, the application of so-called environmental DNA (eDNA) analysis is shown to provide a potent tool for elucidating mechanistic insights in ecological and evolutionary processes (Bohmann et al, 2014). Though the identification of DNA is commonly done by PCR, now LAMP, which can speed up the implementation of management actions, either to protect or eradicate the organism of interest, is on the way (Lee et al, 2017). Generally, in test samples for gene analysis, there are not only viable cells, but also dead cells and extracellular DNA. Thus, to understand clearly what roles they play in the LAMP assay is of great significance for obtaining accurate target information. In this manuscript, we verified that viable cells, dead cells and extracellular DNA could each function as templates in LAMP assays using the Stx1 gene from E. coli as a model. In addition, we performed a series of experiments to determine why viable bacteria could work as templates during LAMP incubation. Inhibitory effects of the complex substances in natural seawater on the analysis sensitivity were also discussed.
2 Materials and methods 2.1 Culture and quantification of E. coliThe strain of E. coli (ATCC 43888) used in this study was purchased from BeNa Culture Collection Co. LTD (Beijing, China). Culture and quantification were performed according to the method we reported previously (Zhang et al, 2018). In brief, once the strain was taken from -80℃, it was pre-grown in liquid LB medium at 37℃ overnight with constant shaking. Next, cultures were inoculated into 200 ml of LB medium and incubated at 37℃ for 8~10 h to achieve mid-exponential phase. Then, the obtained E. coli in the medium was diluted to desired concentrations immediately for further experiments. The densities (CFU/ml) of the un-diluted E. coli cultures were calculated from the averages of colony counts and the magnitude of culture dilution by the plate counting method. Aqueous solutions were prepared with ultrapure water (Resistivity: 18.2 MΩ/cm) produced by a Poseidon-R70 water purification system (Research Scientific Instruments Co. LTD, Xiamen, China).
2.2 Preparation of DNA templates and LAMP primersGenomic DNA templates were prepared according to the method we reported previously (Zhang et al, 2018; 2014). In brief, bacteria in the culture medium were collected by centrifugation. Then the genomic DNA was extracted using a Bacteria Genomic DNA Extraction Kit (Tiangen Biotech Co., LTD, Beijing, China) according to the manufacturer's instructions. The purity was within the acceptable range of 1.8~2.0 A260 nm/A280 nm. The stock concentration of the extracted genomic DNA (36.80 ng/μl) was determined with a Smartspec Plus spectrophotometer (Bio-Rad Lab, USA). Subsequent dilutions at the desired concentrations were stored at 4℃ for no longer than one week prior to use.
Primers for detecting Stx1 gene of E. coli were produced by Beijing SBS Genetech Co., LTD. (Beijing, China) with the following sequences (Table 1). The specificity of these primers has been confirmed previously by Zhao et al (2011) and Yan et al (2017).
|
|
Table 1 Primers used for detecting the Stx1gene of E. coli with the LAMP assay |
Cultured E. coli were filtered with Sterivex cartridges (SVGPL10RC, Millipore, Billerica, MA). Then, concentrated cells were released from the filter by adding 0.9 ml of elution buffer (Stedtfeld et al, 2014). Pure viable cells were obtained by the method of Youn et al (2016) with minor modifications. In brief, 400 μl of bacterial resuspension (108 CFU/ml) and 100 μl of 0.5 mmol/L propidium monoazide (PMA, Biotium Inc., USA) were mixed. After 5 min incubation at room temperature in the dark, the mixtures were light-exposed for 15 min using a PMA-Lite™ LED photolysis device (Biotium Inc., USA) according to the manufacturer's instructions. Before being used as template, the viable cells were carefully washed three times with PBS buffer to remove residual PMA, followed by resuspension with water and density calculation by plate counting. To obtain dead cells, the bacteria in the resuspension were heat-killed by exposure at 95℃ for 10 min (Ahmad et al, 2017). The death and viability of bacteria were confirmed by growth characterization.
2.4 LAMP assaysLAMP assays were performed as previous reported method with minor modifications (Zhang et al, 2014). In brief, a total volume of 25 μl reaction mixture containing 0.2 mmol/L each of F3 and B3, 0.8 mmol/L each of LoopF and LoopB, 1.6 mmol/L each of FIP and BIP, primers, 1.2 mmol/L each of deoxynucleotide triphosphate, 6 mmol/L MgSO4, 1 μl of 10× Bst ThermoPol reaction buffer, 8 U of Bst DNA polymerase large fragment (NEB Co., LTD., USA), and 5 μl of template (viable cells, dead cells, extracted genomic DNA, aqueous samples without pretreatment, etc.), was incubated at 63℃ for 60 min in a thermal cycler (BioRad, Temecula, USA). An electrophoresis apparatus (DY-6, Xinghua Assay Apparatus Factory, Beijing, China) and a DNR bio-imaging systems (MF-ChemiBis 3.2, Israel) were used for electrophoresis analysis with 2.5% agarose gel. DL2000 DNA markers were purchased from TaKaRa Co., LTD (Dalian, China). Moreover, the fluorescent dye GeneFinder (Biov LTD., Xiamen, China) was used for visual characterization of LAMP reaction products. For each measurement, 3.68 pg extracted genomic DNA and ultrapure water were used as templates for positive and negative controls, respectively.
2.5 Spiked experimentsUnpurified tap water samples and natural seawater samples (collected from Jiaozhou Bay, China) were filtered with 0.22 μm Sterivex filters to remove bacteria followed by Silicone membranes (EMD Millipore Corp., Billerica, MA) to remove extracellular DNA (Stedtfeld et al, 2016). Then, bacteria or DNA were added to the filtered water to perform spiked experiments. Unless stated explicitly, all spiked water samples were used within 20 min to prevent the interference resulting cell lysis.
After incubation for ~10 h, a 1000-fold dilution of the initial culture medium was prepared with water, in which the density of E. coli was obtained by the plate counting method. The samples were prepared as follows: 1) To mock real samples, we added 100, 50, 20, 10, 5, 2 and 0 μl of a 1000-fold dilution (containing 109 CFU/ml E. coli) into 19.900, 19.950, 19.980, 19.990, 19.995 and 19.998 ml filtered tap water or seawater. 2) To simulate extracellular DNA in test samples, extracted genomic DNA was added to filtered tap water or seawater at final concentrations of 2.2×100, 2.2×101, 2.2×102, 2.2×103, 2.2×104, 2.2×105 and 0 fg/μl. 3) Bacteria suspensions (1012 CFU/ml) of pure viable cells and dead cells were added into filtered tap water or seawater for final concentrations ranging from 0 to 107 CFU/ml.
2.6 Detection of the Stx1 gene in natural seawaterReal seawater samples collected from Shilaoren Beach of Qingdao (120°28.308'E; 36°5.502'N) were transferred to the lab at ~8℃ and were used immediately. The Stx1 genes in the samples were assayed by LAMP via three approaches: 1) Viable bacteria were collected as described above and used as template; 2) Five microliters of unpurified seawater sample were directly added to the reaction mixture; 3) After boiling treatment, 5 μl of the seawater sample was added into the reaction mixture. Note, in both of the second and third approach, 0.4×Bst ThermoPol Reaction Buffer was used for LAMP to lower the final ionic strength.
2.7 Characterization of the survival behavior of viable cells during LAMP incubation at 63℃An electrical bacterial growth sensor was used to characterize the survival behavior of E. coli according to our previously published method (Zhang et al, 2018). First, to monitor bacterial growth, each disposable glass reaction tube was loaded with 1.3 ml sterile LB medium. Next, 20 μl of a suspension (containing 107 CFU of pure viable cells) was added into the LAMP reaction mixture (200 μl in total), followed by incubation at 63℃. During the incubation, 10 μl of reaction mixture was transferred into a disposable glass reaction tube at 0, 1, 2, 3, 4 and 5 min, respectively. Disposable glass reaction tubes were simultaneously inserted into working channels of the electrical bacterial growth sensor. Apparent conductivity values were collected with the excitation frequency of 2.0 MHz, excitation amplitude of 16 V and recording rate of 10 second, and were used to generate growth curves by plotting against incubation time. Because the critical factor was to monitor conductivity changes during the process of bacterial growth rather than determination of the absolute value, an algorithm is presented here to obtain normalized apparent conductivity value (NACV), which was calculated as follows:
| $ \text{NAC}{{\text{V}}_{\text{n}}}\text{=AC}{{\text{V}}_{\text{n}}}\text{-AC}{{\text{V}}_{\text{0}}} $ | (1) |
where NACVn was the real time normalized apparent conductivity value of bacterial culture medium in the tube obtained at point n; ACVn was the apparent conductivity value collected at point n; ACV0 was the first apparent conductivity value collected at the beginning of the incubation. Namely, the apparent conductivity value at time = 0 was subtracted from all the following data during monitoring in order to normalize the starting growth levels at zero for all conditions. In addition, the survival behavior of E. coli was characterized by the plate culture method for comparison.
3 Results 3.1 LAMP assay of the Stx1 gene in mock samplesAfter an incubation of ~10 h, the density of E. coli in the culture medium was 1.83×1012 CFU/ml, as confirmed by the plate counting method. The following steps were used to prepare templates for the LAMP assays: 1) A 1000-fold dilution of the initial culture medium was prepared. 2) After the initial culture medium was passed through a 0.22 μm filter to remove bacteria and other large molecules, a 1000-fold dilution of the filtrate was prepared. 3) The filtrate (400 μl) was treated with PMA to make extracellular DNA non-amplifiable (Youn et al, 2016; Ahmad et al, 2017; Nocker et al, 2006). 4) According to the method reported by Youn et al(2016), pure viable cells were obtained by eliminating amplifiable gene fragments from dead cells and extracellular DNA, and then they were re-suspended with water to prepare a bacterial suspension (107 CFU/ml). 5) Half of the viable bacterial suspension was heated to prepare a dead cell suspension.
Five microliters of these five template preparations were added into respective reaction mixtures, and the LAMP assay was performed simultaneously in triplicate. The results were characterized by gel electrophoresis and the fluorescent dye GeneFinder, which has been proven to yield accurate quantification (Zhang et al, 2011). As shown in Fig. 1, all of the amplification reactions were successful, with the exception of one with PMA-treated supernatant from diluted culture medium. Furthermore, the results of positive and negative control experiments indicated that the reaction system was reliable.

|
Figure 1 Detection of the Stx1 gene by LAMP assay with different templates Results were determined by gel electrophoresis (a) and the fluorescent dye GeneFinder (b). In the positive control, 3.68 pg extracted genomic DNA was used as template, and in the negative control, pure water was used as template |
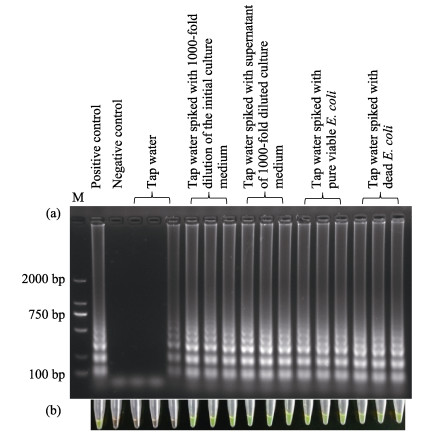
After a ~8 h incubation, a 1000-fold dilution of the initial culture medium, in which the density of E. coli was 9.95×108 CFU/ml, was prepared. Then it was used to spike tap water samples as follows: 1) In the first group, no E. coli was added. 2) In the second group, 50 μl of the 1000-fold dilution were added into 19.950 ml tap water. 3) Cells and other large molecules in 50 μl of 1000-fold dilution were removed by filtering. Then the filtrate was added to 19.950 ml tap water. 4) Pure viable E. coli in 50 μl of the 1000-fold dilution were obtained via filtering and PMA-treatment, followed by addition to 19.950 ml tap water. 5) Pure viable E. coli in another 50 μl of 1000-fold dilution was killed by heating, also followed by addition to 19.950 ml tap water.
Five microliters of these five preparations were used as a template for each LAMP assay reaction. The gel electrophoresis and visualization results are shown in Fig. 2. The Stx1 gene could clearly be detected in all spiked samples. Note, however, that there were not universal color changes in all three reaction tubes characterized with Genefinder, and that a ladder-like pattern of amplicons was present on one of the gel lanes. This phenomenon indicates that ⅰ) the sensitivity of the gel electrophoresis was higher than the sensitivity of the visual assay; and ⅱ) there was a small amount of extracellular DNA in the unpurified tap water.
|
Figure 2 Detection of the Stx1 gene in spiked tap water samples by LAMP assay Results were determined by gel electrophoresis (a) and the fluorescent dye GeneFinder (b). The positive and negative controls were the same in Fig. 1. Pure water and 3.68 pg extracted genomic DNA were used as negative and positive template, respectively, in control experiments |
Twenty milliliters of tap water samples were spiked with 1.99×104, 4.98×104, 9.95×104, 1.99×105, 4.98×105 or 9.95×105 CFU E. coli in culture medium. When 5 μl of the spiked samples were used as a template for the LAMP assay, the Stx1 gene could be detected in all reactions, characterized both by gel electrophoresis and the fluorescent dye GeneFinder. The Stx1 gene could also be detected in the tap water samples, in which the final concentration of spiked genomic DNA ranged from 2.2×100 to 2.2×105 fg/μl. When tap water samples were spiked with pure viable bacteria at different concentrations, the Stx1 gene was detectable (100%) in the case of ≥9.95×102 CFU/ml, as characterized by gel electrophoresis.
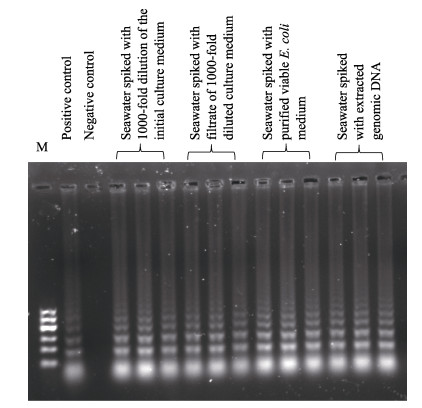
After another incubation of ~8 h, a 1000-fold dilution of the initial culture medium was prepared in water, in which the density of the E. coli was 1.07× 107 CFU/ml. Then, it was used to spike seawater samples as follows: 1) In the first group, 50 μl of 1000- fold dilution were added to 19.950 ml clean seawater. 2) Cells and other large molecules in 50 μl of 1000-fold dilution were removed by filtering. Then the filtrate was added to 19.950 ml filtered seawater. 3) Pure viable E. coli in 50 μl of 1000-fold dilution were obtained via filtering and PMA-treatment, followed by addition into 19.950 ml of filtered seawater. 4) The extracted genomic DNA was added into filtered seawater, allowing a final concentration of 2.2×105 fg/μl.
Five microliters of these five solutions were used as template for the LAMP assay. The gel electrophoresis results are shown in Fig. 3. The Stx1 gene could be detected in all spiked seawater samples. Twenty milliliters of filtered seawater samples were spiked with 2.14×104, 5.35×104, 1.07×105, 2.14×105, 5.35×105 and 1.07×106 CFU E. coli in culture media. When 5 μl of the spiked samples were used as template for the LAMP assay, the Stx1 gene could be detected in all reactions, as characterized by gel electrophoresis. The Stx1 gene could also be detected in all of the spiked seawater samples in which the final concentration of genomic DNA was over the range of 2.2×102~2.2×105 fg/μl. When seawater samples were spiked with pure viable bacteria at different concentration, the Stx1 gene in 5 μl was detectable in all cases (100%) with ≥5.35× 103 CFU/ml, as characterized by gel electrophoresis.
|
Figure 3 Detection of the Stx1 gene in spiked seawater samples by LAMP assay characterizing by gel electrophoresis Pure water and 3.68 pg extracted genomic DNA were used as negative and positive template, respectively, in control experiments |
The density of viable E. coli in the real seawater was calculated to be 19.20 CFU/ml after membrane filtration by the plate counting method. Pure viable bacteria in 100 ml of real seawater were obtained via filtering and PMA-treatment, followed by resuspension in 100 μl PBS buffer. Then, 5 μl of the suspension was used as template for the LAMP assay to detect the Stx1 gene. As shown in Fig. 4 Lane 3~5, the ladder-like pattern of the amplicon was clear. Furthermore, when 5 μl of the natural seawater was directly used as template, with or without boiling-treatment, the Stx1 gene could also be detected (Fig. 4 Lane 6~11). It is worth noting that using undiluted seawater directly as template lowered the sensitivity of the LAMP assay remarkably, perhaps resulting from the inhibition caused by the salinity or complex matrices. We found that 0.4×Bst ThermoPol Reaction Buffer worked well for the amplification task in this case. However, this parameter was not optimized further.
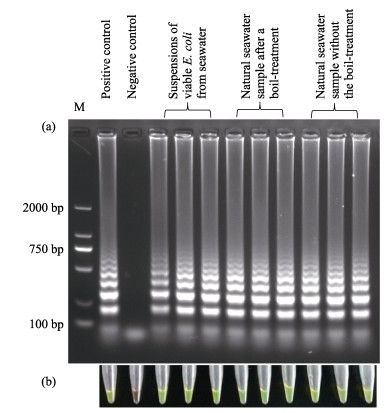
|
Figure 4 Detection of the Stx1 gene in real seawater samples by LAMP assay using templates obtained with different pretreatment methods Pure water and 3.68 pg extracted genomic DNA were used as negative and positive template, respectively, in control experiments |
Using the electrical bacterial growth sensor, we obtained typical curves of E. coli growth (Fig. 5a), which were generated by plotting the NACV against incubation time. When 10 μl of LAMP reaction mixture (containing 104 CFU viable cells) without incubating at 63℃ was transferred into a disposable glass reaction tube for culture at 37℃ in the electrical bacterial growth sensor, there was an S-shaped growth curve. On the curve there was a lag phase, resulting from the stress that bacteria might experience after dilution and/or loading (Settu et al, 2015), as well as the time required for generating enough end products to produce detectable increasing conductivity. The lag phase was followed by an acceleration phase, during which the growth rate increased until a constant growth rate was achieved. Then, an exponential-like phase appeared, followed by a deceleration phase. These results reflect the typical growth of bacteria (Zhang et al, 2018), suggesting adequate vitality of the cells. The rest of the LAMP reaction mixture was incubated at 63℃. At the end of 1, 2, 3, 4 and 5 min, 10 μl of the mixture was transferred into disposable glass reaction tubes for culture at 37℃. A similar S-shaped growth curve was obtained when the inoculum was collected at the end of 1 min incubation, with a significantly increased time of detection (~600 min) compared to that of the inoculum that was collected before incubation at 63℃. This implies that there were still viable cells in the culture medium; nevertheless, the number was much less than that sampled from the LAMP mixture before incubation (Zhang et al, 2018). If 10 μl of inoculum was sampled from the LMAP mixture with incubation at 63℃ for more than 1 min, a horizontal line was obtained due to unchanged conductivity, indicating a nonoccurrence of bacterial growth. In other words, the cells all died completely. The survival behavior of E. coli was also characterized by the plate counting method (Fig. 5b). The results were in good agreement with those obtained with the electrical bacterial growth sensor.

|
Figure 5 Growth characterization for studying survival behavior of viable cells
(a) Growth curves of E. coli (NACV vs. incubation time) in LB medium obtained with the electrical bacterial growth sensor. For the S-shaped curve in red, the inoculum was 10 μl of LAMP reaction mixture (containing 104 CFU viable cells), which was sampled before incubation at 63℃. For the S-shaped curve in blue, the inoculum was 10 μl of the LAMP reaction mixture, which was sampled after 1 min incubation at 63℃. For the other horizontal lines, the inoculum was 10 μl of the LAMP reaction mixture sampled at the end of 2, 3, 4 and 5 min incubation. In each disposable glass reaction tube 1.3 ml LB medium was loaded. The operation parameters were as follows: excitation frequency of 2.0 MHz; excitation amplitude of 16 V; recording rate per 10 s. (b) Photographs of E. coli colonies. For plates number 1 to 6 the inoculum was 10 μl of LAMP reaction mixture containing viable cells, which was sampled at the end of 0, 1, 2, 3, 4 and 5 min of incubation at 63℃, respectively |
The detection of target genes with PCR techniques commonly requires cell lysis, DNA extraction and template purification; necessitates the use of bulky instrumentations; and remains cumbersome (Stedtfeld et al, 2014; Enomoto et al, 2005). LAMP has the potential to circumvent these issues because of the reduced dependency on pretreatment of the test samples (Safavieh et al, 2016; Njiru et al, 2012; Williams et al, 2017), lowering the need for highly sophisticated equipment and turnaround times (Zhao et al, 2015; Zhang et al, 2014). However, to avoid false negatives and overestimation of viable cell numbers when performing LAMP, greater understanding of the role of templates is still needed at present.
We found that a 1000-fold diluted filtrate of culture medium in which cells had been removed(Kanitkar et al, 2017; Stedtfeld et al, 2016), could be used as template for the LAMP assay to detect the Stx1 gene, indicating that in this case 1) there was extracellular DNA of E. coli in the filtrate, as a result of natural lysis of dead cells or active secretion of viable cells (Biller et al, 2017); and 2) the reaction system was tolerant to the substances introduced by the culture medium. Upon exposure to blue light, PMA can intercalate into double stranded DNA and form covalent linkages, resulting in chemically modified DNA, which cannot be amplified by PCR or LAMP reactions (Youn et al, 2016; Nocker et al, 2006; Chen et al, 2011). Therefore, when this template, i.e. the 1000-fold diluted filtrate of culture medium, was treated with PMA, the Stx1 gene could not be detected by the LAMP assay. This suggested that in this case all of the extracellular DNA were non-amplifiable. We considered the adequate PMA making the difference from the phenomenon found by Ahmad et al (2017).
Our results confirmed that in culture medium at exponential phase there were both viable cells and extracellular DNA. Pure viable cells, dead cells, extracellular DNA and their mixture in culture medium could each be used as templates for the LAMP assay, reconfirming the non-ignorable effects of coexisting extracellular DNA in determining viable cells by DNA-based molecular detection techniques (Ahmad et al, 2017; Chen et al, 2011). Even when all of the cells were removed by filtration, the Stx1 gene in tap water could be sensed by the LAMP assay as determined by gel electrophoresis. Using DNA extraction-free LAMP for detecting the vcrA gene in groundwater samples, Stedtfeld et al (2014) found that filtration of larger volumes caused loss of sensitivity, which they attributed to the loss of extracellular DNA that could pass through the cartridge. Our results confirmed this hypothesis.
The results of spiked experiments also showed that the target gene in pure viable cells, dead cells and extracellular DNA could be detected by the LAMP assay, whether it was from tap water or seawater. While the detectable concentrations of spiked genomic DNA in tap water and seawater were over the range of 2.2×100~2.2×105 fg/μl and 2.2×102~2.2×105 fg/μl, respectively, the detection limits of pure viable bacteria in tap water and seawater were 9.95×102 CFU/ml and 5.35×103 CFU/ml, respectively, suggesting that substances in seawater showed considerable inhibitory effects on LAMP reactions, as they did in the culture medium (Enomoto et al, 2005) and other real and mock samples (Stedtfeld et al, 2014; Feng et al, 2018). Using 5 μl of seawater as template in which the concentration of spiked pure viable bacteria was 5.35×100 CFU/ml, we did not have successful amplification. However, when the cells from 100 ml of this spiked seawater were collected by filtering, followed by resuspension, the target gene could be detected clearly, suggesting that a step of enrichment by filtering (Stedtfeld et al, 2014; Stedtfeld et al, 2016) or centrifugation (Kanitkar et al, 2017) was an effective approach for detecting target genes at low content samples, while omission of DNA extraction could save both time and labor in preparing templates for the LAMP assay.
Among the reports of direct LAMP, performed without DNA extraction and purification steps, heat-treatment of samples was considered to benefit the reactions due to the increased cell lysis (Poon et al, 2006; Njiru et al, 2008; Ihira et al, 2010). However, Stedtfeld et al (2016) as well as Ahmad et al (2017) found that heat-treatment of samples prior to LAMP had no observable influence on the time at which the SNR reached an arbitrary cut-off of 10 (Tt) or sensitivity compared to direct amplification without lysis. In particular, viable cells could be used as template directly. There had been various hypotheses for how they work. Ahmad et al (2017) considered that the success of the LAMP assay might be due to increased cell permeability at a temperature of 63℃, providing access to the genetic contents in the reaction solution. Stedtfeld et al (2014) thought that perhaps the smaller Bst polymerase had a high rate of passage into intact bacterial cells, or maybe cells become porous at the amplification reaction temperature of 63℃. Kanitkar et al (2017) hypothesized that cell lysis occurred while incubating LAMP reactions at 63℃, though they didnxt think the lysis was adequate. Until now, the mechanisms of unintentional lysis were unclear. The results of our survival experiments showed that the number of viable cells in the LAMP reaction mixture only slightly decreased after incubation at room temperature for 10 min but decreased sharply during incubation at 63℃. All of the viable cells in the LAMP mixture died completely when the incubation was more than 2 min. However, in control experiments, in which water was used in place of the LAMP mixture, more than half of the spiked viable E. coli survived at the end of 10 min. These results confirmed that viable cells were killed rapidly in the LAMP reaction mixture during the incubation at 63℃. This discovery explained the moderate influence on Tt and the sensitivity of heat-treatment prior to LAMP (Stedtfeld et al, 2016; Ahmad et al, 2017). But it was unclear now which species were at work for the killing.
Shipping of samples to an off-site laboratory is cumbersome, costly, and requires proper disposal, which influences the number of replicates that are generally analyzed. Transportation of samples also causes chances for leaking/contamination, and increased storage time has the potential to alter the integrity of unstable biomarkers (Stedtfeld et al. 2014). Thus, the validation of the LAMP assay, which can be performed at the point of care due to the ability to use test samples directly as template, has potential benefits for multiple researchers. In particular, the application of eDNA analysis for ecological research, such as the distribution of aquatic and terrestrial organisms and their biodiversity, is becoming increasingly more popular (Williams et al, 2017; Bohmann et al, 2014; Kristy et al, 2016; Shan et al, 2018). The combination of eDNA with direct LAMP may provide a beneficial combination. From our results using spiked experiments and the detection of the Stx1 gene in real seawater samples, we could see that the omission of DNA extraction and purification saved both time and labor (Enomoto et al, 2005). However, this was achieved at the expense of sensitivity and accuracy, due to variation in the extracellular DNA, as well as inhibitory factors. Furthermore, quantitative correlation between the copy number of marker genes in eDNA and the number of target organisms commonly is unstable (Williams et al, 2017). Therefore, to design a strategy for field-based analysis, these factors are worthy of additional consideration.
We confirmed that viable cells, dead cells and extracellular DNA all can function as template for the LAMP assay, using the Stx1 gene of E. coli as a model. Thus, to detect similar target bacteria in samples, extracellular DNA could potentially be captured with silica membranes (Harikai et al, 2015) as an alternative to collecting cells by filtration (Stedtfeld et al, 2014). Moreover, in the case of quantifying viable cells, before determining target genes with LAMP, analysts should take additional steps, such as PMA-treatment, to eliminate interference from co-existing extracellular DNA. Otherwise the quantities might be overestimated.
Samples (diluted culture medium, tap water and seawater) can be directly used as template for the LAMP assay without physical, chemical or biochemical pretreatment, due to the presence of viable cells, dead cells or extracellular DNA. Direct amplification from unpurified samples could reduce the complexity of the gene analysis instruments, as well as difficulties of operation, allowing for simple, in situ, rapid and cost- effective gene analysis. However, common pretreatment approaches, e.g., concentration and purification, are useful for higher sensitivity and accuracy. This should be considered carefully when designing experiments for detecting target bacteria in specific samples, especially at low concentration. In addition, when researchers combine LAMP and eDNA, the quantitative correlation between the copy number of marker genes in eDNA and the target number of organisms should be closely considered.
5 ConclusionUsing the Stx1 gene from E. coli as model, we verified that viable cells, dead cells and extracellular DNA could function as template in the LAMP assay. The target gene in viable E. coli can be amplified due to the efficient lysis of cells in the LAMP reaction mixture during the incubation at 63℃, allowing for replication of the target nucleic acid in the presence of Bst DNA polymerase. Therefore, heat-treatment is not necessary prior to LAMP assay. In addition, the Stx1 gene in diluted culture medium, spiked tap water, spiked seawater and real seawater all could be detected, with or without the step of DNA extraction. However, the complex substances in real sample (e.g. natural seawater) exhibited considerable inhibitory effect on the sensitivity of the LAMP assay. These outcomes are meaningful for building a point-of-care strategy by employing the LAMP assay for environmental monitoring, bio-resource surveys, food safety, etc. in particular those based on environmental DNA.
Conflict of interestThe authors declare that they have no conflict of interest.
| Abdul-Ghani R, Al-Mekhlafi AM, Karanis P. Loop-mediated isothermal amplification (LAMP) for malarial parasites of humans: Would it come to clinical reality as a point-of-care test?. Acta Tropica, 2012, 122(3): 233-240 DOI:10.1016/j.actatropica.2012.02.004 | |
| Ahmad F, Stedtfeld RD, Waseem H, et al. Most probable number-loop mediated isothermal amplification (MPN-LAMP) for quantifying waterborne pathogens in < 25 min. Journal of Microbiological Methods, 2017, 132: 27-33 DOI:10.1016/j.mimet.2016.11.010 | |
| Bektas A, Chapela I. Efficiency of a fluorescent, non-extraction LAMP DNA amplification method: Toward a field-based specific detection of maize pollen grains. Aerobiologia, 2016, 32(3): 481-488 DOI:10.1007/s10453-016-9420-z | |
| Biller SJ, Mcdaniel LD, Breitbart M, et al. Membrane vesicles in sea water: Heterogeneous DNA content and implications for viral abundance estimates. ISME Journal, 2017, 11(2): 394-404 DOI:10.1038/ismej.2016.134 | |
| Bohmann K, Evans A, Gilbert MT, et al. Environmental DNA for wildlife biology and biodiversity monitoring. Trends in Ecology & Evolution, 2014, 29(6): 358-367 | |
| Chen S, Wang F, Beaulieu JC, et al. Rapid detection of viable salmonellae in produce by coupling propidium monoazide with loop-mediated isothermal amplification. Applied and Environmental Microbiology, 2011, 77(12): 4008-4016 DOI:10.1128/AEM.00354-11 | |
| Enomoto Y, Yoshikawa T, Ihira M, et al. Rapid diagnosis of herpes simplex virus infection by a loop-mediated isothermal amplification method. Journal of Clinical Microbiology, 2005, 43(2): 951-955 DOI:10.1128/JCM.43.2.951-955.2005 | |
| Feng N, Zhou Y, Fan Y, et al. Yersinia pestisdetection by loop-mediated isothermal amplification combined with magnetic bead capture of DNA. Brazilian Journal of Microbiology, 2018, 49(1): 128-137 DOI:10.1016/j.bjm.2017.03.014 | |
| Giovannoni SJ, Britschgi TB, Moyer CL, et al. Genetic diversity in Sargasso Sea bacterioplankton. Nature, 1990, 345: 60-63 DOI:10.1038/345060a0 | |
| Goda T, Tabata M, Miyahara Y. Electrical and electrochemical monitoring of nucleic acid amplification. Frontiers in Bioengineering and Biotechnology, 2015, 3: 29 | |
| Harikai N, Shinomiya K. Application of an alkaline and silica membrane DNA extraction method to detect mitochondrial DNA in foods. Food Analytical Methods, 2015, 8(5): 1215-1224 DOI:10.1007/s12161-014-0002-9 | |
| Hayashida K, Kajino K, Hachaambwa L, et al. Direct blood dry LAMP: A rapid, stable, and easy diagnostic tool for human African trypanosomiasis. PLoS Neglected Tropical Diseases, 2015, 9(3): e0003578 DOI:10.1371/journal.pntd.0003578 | |
| Hill J, Beriwal S, Chandra I, et al. Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. Journal of Clinical Microbiology, 2008, 46(8): 2800 DOI:10.1128/JCM.00152-08 | |
| Ihira M, Sugiyama H, Enomoto Y, et al. Direct detection of human herpesvirus 6 DNA in serum by variant specific loop-mediated isothermal amplification in hematopoietic stem cell transplant recipients. Journal of Virological Methods, 2010, 167(1): 103-106 DOI:10.1016/j.jviromet.2010.01.025 | |
| Kaneko H, Kawana T, Fukushima E, et al. Tolerace of loop-mediated isothermal amplification to a culture medium and biological substances. Journal of Biochemical and Biophysical Methods, 2007, 70(3): 499-501 DOI:10.1016/j.jbbm.2006.08.008 | |
| Kanitkar YH, Stedtfeld RD, Hatzinger PB, et al. Development and application of a rapid, user-friendly, and inexpensive method to detect Dehalococcoides sp. reductive dehalogenase genes from groundwater. Applied Microbiology and Biotechnology, 2017, 101: 4827-4835 DOI:10.1007/s00253-017-8203-y | |
| Kiddle G, Hardinge P, Buttigieg N, et al. GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnology, 2012, 12(1): 15 | |
| Koizumi N, Nakajima C, Harunari T, et al. A new loop-mediated isothermal amplification method for rapid, simple, and sensitive detection of Leptospira spp. in urine. Journal of Clinical Microbiology, 2012, 50(6): 2072-2074 DOI:10.1128/JCM.00481-12 | |
| Kristy D, Fronhofer EA, Elvira M, et al. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nature Communications, 2016, 7: 12544 DOI:10.1038/ncomms12544 | |
| Lee D, Kim YT, Lee JW, et al. An integrated direct loop-mediated isothermal amplification microdevice incorporated with an immunochromatographic strip for bacteria detection in human whole blood and milk without a sample preparation step. Biosensors and Bioelectronics, 2016, 79: 273-279 DOI:10.1016/j.bios.2015.12.044 | |
| Lee PL. DNA amplification in the field: Move over PCR, here comes LAMP. Molecular Ecology Resources, 2017, 17(2): 138-141 DOI:10.1111/1755-0998.12548 | |
| Njiru ZK, et al. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Neglected Tropical Diseases, 2008, 2: e147 DOI:10.1371/journal.pntd.0000147 | |
| Njiru ZK. Loop-mediated isothermal amplification technology: Towards point of care diagnostics. PLoS Neglected Tropical Diseases, 2012, 6(6): e1572 DOI:10.1371/journal.pntd.0001572 | |
| Nocker A, Cheung CY, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. Journal of Microbiological Methods, 2006, 67(2): 310-320 DOI:10.1016/j.mimet.2006.04.015 | |
| Patterson AS, Heithoff DM, Ferguson BS, et al. Microfluidic chip-based detection and intraspecies strain discrimination of salmonella serovars derived from whole blood of septic mice. Applied and Environmental Microbiology, 2013, 79(7): 2302-2311 DOI:10.1128/AEM.03882-12 | |
| Poon LL, Wong BW, Ma EH, et al. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clinical Chemistry, 2006, 52(2): 303-306 DOI:10.1373/clinchem.2005.057901 | |
| Priye A, Bird SW, Light YK, et al. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Scientific Reports, 2017, 7: 44778 DOI:10.1038/srep44778 | |
| Safavieh M, Kanakasabapathy MK, Tarlan F, et al. Emerging loop-mediated isothermal amplification-based microchip and microdevice technologies for nucleic acid detection. ACS Biomaterials Science & Engineering, 2016, 2(3): 278 | |
| Settu K, Chen CJ, Liu JT, et al. Impedimetric method for measuring ultra-low E. coli concentrations in human urine. Biosensors and Bioelectronics, 2015, 66: 244-250 DOI:10.1016/j.bios.2014.11.027 | |
| Shan X, Li M, Wang W. Application of environmental DNA technology in aquatic ecosystem. Progress in Fishery Sciences, 2018, 39(3): 23-29 | |
| Soejima M, Egashira K, Kawano H, et al. Rapid detection of haptoglobin gene deletion in alkaline-denatured blood by loop-mediated isothermal amplification reaction. Journal of Molecular Diagnostics, 2011, 13(3): 334-339 DOI:10.1016/j.jmoldx.2011.01.005 | |
| Stedtfeld R, Stedtfeld TM, Kronlein M, et al. DNA extraction- free quantification of Dehalococcoides spp. in groundwater using a hand-held device. Environmental Science & Technology, 2014, 48(23): 13855 | |
| Stedtfeld RD, Stedtfeld TM, Samhan F, et al. Direct loop mediated isothermal amplification on filters for quantification of Dehalobacter in groundwater. Journal of Microbiological Methods, 2016, 131: 61-67 DOI:10.1016/j.mimet.2016.09.025 | |
| Stoeck T, Bass D, Nebel M, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Molecular Ecology, 2010, 19: 21-31 DOI:10.1111/j.1365-294X.2009.04480.x | |
| Wang L, Shi L, Alam MJ, et al. Specific and rapid detection of foodborne Salmonella by loop-mediated isothermal amplification method. Food Research International, 2008, 41(1): 69-74 DOI:10.1016/j.foodres.2007.09.005 | |
| Williams MR, Stedtfeld RD, Engle C, et al. Isothermal amplification of environmental DNA (eDNA) for direct field-based monitoring and laboratory confirmation of Dreissena sp. PLoS One, 2017, 12(10): e0186462 DOI:10.1371/journal.pone.0186462 | |
| Williams MR, Stedtfeld RD, Waseem H, et al. Implications of direct amplification for measuring antimicrobial resistance using point-of-care devices. Analytical Methods, 2017, 9(8): 1229-1241 DOI:10.1039/C6AY03405E | |
| Yan M, Li W, Zhou Z, et al. Direct detection of various pathogens by loop-mediated isothermal amplification assays on bacterial culture and bacterial colony. Microbial Pathogenesis, 2017, 102: 1-7 DOI:10.1016/j.micpath.2016.10.025 | |
| Youn SY, Jeong OM, Choi BK, et al. Application of loop-mediated isothermal amplification with propidium monoazide treatment to detect live Salmonella in chicken carcasses. Poultry Science, 2016, 96: 458-464 | |
| Zhao X, Li Y, Wang L, et al. Development and application of a loop-mediated isothermal amplification method on rapid detection Escherichia coli O157 strains from food samples. Food & Fermentation Industries, 2011, 37(5): 2183-2188 | |
| Zhao Y, Chen F, Li Q, et al. Isothermal amplification of nucleic acids. Chemical Reviews, 2015, 115(22): 12491 DOI:10.1021/acs.chemrev.5b00428 | |
| Zhang X, Liu W, Lu X, et al. Monitoring the progression of loop-mediated isothermal amplification using conductivity. Analytical Biochemistry, 2014, 466: 16-18 DOI:10.1016/j.ab.2014.08.002 | |
| Zhang X, Lowe SB, Gooding JJ. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosensors and Bioelectronics, 2014, 61(20): 491-499 | |
| Zhang X, Qu K, Li Q, et al. Recording the reaction process of loop-mediated isothermal amplification (LAMP) by monitoring the voltammetric response of 2x-deoxyguanosine 5x-triphosphate. Electroanalysis, 2011, 23(10): 2438-2445 DOI:10.1002/elan.201100279 | |
| Zhang X, Jiang X, Yang Q, et al. Online monitoring of bacterial growth with electrical sensor. Analytical Chemistry, 2018, 90(10): 6006-6011 DOI:10.1021/acs.analchem.8b01214 | |
| Zielińska MS, Kidawa DD, Stempniewicz PL, et al. Environmental DNA as a valuable and unique source of information about ecological networks in Arctic terrestrial ecosystems. Environmental Reviews, 2017, 25: 282-291 DOI:10.1139/er-2016-0060 |



